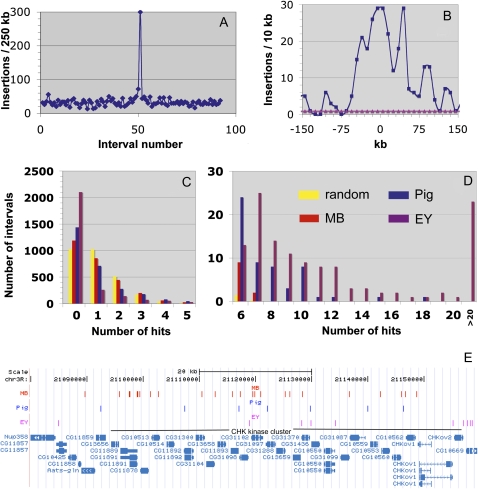
Figure 2 .
Saturation behavior of P, piggyBac, and Minos insertions. (A) Plot of MB insertions per 250 kb vs. interval number along chromosome 3L reveals a large hotspot. (B) MB insertions within 10-kb intervals around the hotspot in A. The number per interval expected by chance is shown in pink. 0 corresponds to 3L:12580233, the site on the homolog of the mobilized element in the MB screen. (C and D) Distribution of MB (red), piggyBac (blue), or EY (purple) insertions within 10-kb genomic intervals on chromosome 3R, compared with random transposition (Poisson distribution, yellow). To facilitate comparison, the same numbers of insertions were analyzed in each case (2790; corresponding to 1 insertion per interval). The number of intervals with 0 insertions (C, “0”) is relevant to coldspot behavior; intervals hit more frequently than by random expectation (D) are indicative of piggyBac and P-element hotspots. (E) The Minos hotspot located within a cluster of genes encoding CHK kinases on chromosome 3R. The locations of MB (Minos), Pig (piggyBac), and EY (P) element insertions are shown by vertical bars above the gene map of the region.