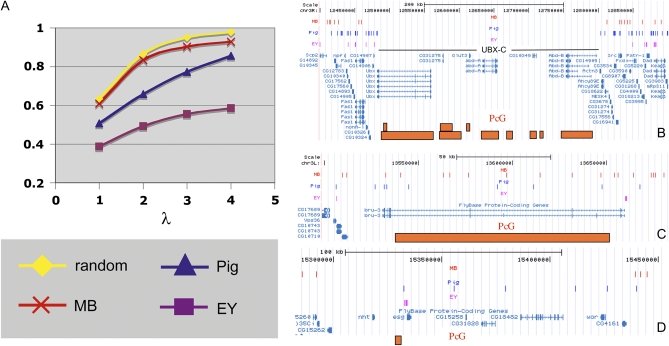
Figure 4 .
Transposons nonrandomly avoid some genomic intervals, including regions with PcG-dependent repressive marks. (A) The saturation behavior of 40-kb genomic intervals for transposon insertion on chromosome 3R is plotted as λ (the ratio of number of insertions/number of intervals) increases. Poisson (random) expectation (yellow), MB (Minos) elements (red), piggyBac elements (blue), and EY (P) elements (purple). EY elements saturate well below 100%. In contrast, MB elements approach saturation only slightly more slowly than random, whereas piggyBacs appear intermediate. (B) MB, piggyBac, and EY elements insert with greatly reduced frequency in the Bithorax gene cluster. Regions of the Drosophila genome as displayed on the UCSC browser are shown. Insertion sites for these elements are shown in labeled tracks above the map as vertical lines of unit thickness (MB in red; piggyBac in blue; EY in purple; thicker lines denote multiple insertions). The orange boxes display the approximate position of PcG-target regions as mapped by Schwartz et al. (2010). (C) Similar display of the bru-3 gene region shows that not all Polycomb-regulated chromatin domains are transposon poor. (D) The esg gene cluster and its surrounding region illustrates that some PcG targets are largely refractory to MB insertion, but not to the other two elements.