Abstract
The fate of a newly arising beneficial mutation depends on many factors, such as the population size and the availability and fitness effects of other mutations that accumulate in the population. It has proved difficult to understand how these factors influence the trajectories of particular mutations, since experiments have primarily focused on characterizing successful clones emerging from a small number of evolving populations. Here, we present the results of a massively parallel experiment designed to measure the full spectrum of possible fates of new beneficial mutations in hundreds of experimental yeast populations, whether these mutations are ultimately successful or not. Using strains in which a particular class of beneficial mutation is detectable by fluorescence, we followed the trajectories of these beneficial mutations across 592 independent populations for 1000 generations. We find that the fitness advantage provided by individual mutations plays a surprisingly small role. Rather, underlying “background” genetic variation is quickly generated in our initially clonal populations and plays a crucial role in determining the fate of each individual beneficial mutation in the evolving population.
THE simplest models of adaptation assume that mutations are rare, so the fate of each beneficial mutation is decided on its own merits: it increases in frequency (or is lost due to random drift) at a rate commensurate with its selective advantage (Ewens 2004). Recent experiments, however, have shown that even for modestly sized populations, beneficial mutation rates are large enough that multiple mutations will often interfere or spread simultaneously [this is referred to as “clonal interference” or “multiple mutation effects” (De Visser et al. 1999; Miralles et al. 1999; Joseph and Hall 2004; De Visser and Rozen 2006; Desai et al. 2007; Gresham et al. 2008; Kao and Sherlock 2008; Miller et al. 2011)].
In sexual populations, if recombination is sufficiently common it can break up the linkage between mutations and ensure that each is selected independently (Peters and Otto 2003). But in asexual populations, or on genomic distance scales in sexual organisms where recombination is sufficiently rare, the fates of mutations are intertwined. This means that the fate of each beneficial mutation depends on many parameters: mutation rate, population size, and the distribution of fitness effects all control the availability of competing mutations that can help or hinder the spread of each new beneficial mutation (Gerrish and Lenski 1998; Wilke 2004; Desai and Fisher 2007; Fogle et al. 2008; Rouzine et al. 2008). Despite extensive theoretical work, we still have no way of predicting how the fate of individual mutations should depend on all these factors (see Park et al. 2010 for a recent review).
It has also proved difficult to measure experimentally how these effects determine the fate of individual mutations, although a variety of experimental approaches have provided insight into these dynamics (see Elena and Lenski 2003; De Visser and Rozen 2006; Hegreness and Kishony 2007; Buckling et al. 2009 for reviews). Some of this work has inferred the presence of interference effects by observing how adaptation rates depend on population sizes and mutation rates (De Visser et al. 1999; Desai et al. 2007). This helps us understand the overall effects of clonal interference and multiple-mutation effects, but still falls well short of a direct and detailed description of mutation dynamics in evolving populations. Other experiments have relied on genetically labeling different subsets of the population at the start of an experiment and inferring interference between mutations on the basis of the increase or decrease in the frequency of mutations with particular labels (Hegreness et al. 1996; Kao and Sherlock 2008). This kind of experiment provides some insight into the dynamics of mutations, but does not allow us to observe the fates of individual mutations, and focuses only on those that are successful enough to influence marker frequencies. Other work has used sequencing of evolving populations to find specific examples of clonal interference and multiple-mutation effects (Bollback and Huelsenbeck 2007; Gresham et al. 2008; Barrick and Lenski 2009; Barrick et al. 2009; Betancourt 2009). This approach provides a very high-resolution view of the dynamics of individual mutations, leading to substantial insight. For example, a recent study by Miller et al. (2011) used extensive sequencing of adapting phage populations to distinguish between a variety of theoretical models for how interference dynamics determine the fate of beneficial mutations. However, sequencing limitations mean that such studies have generally been restricted to retrospective analysis of relatively few successful clones emerging from a small number of evolving lines and hence have lacked statistical power to investigate how particular factors influence the fate of beneficial mutations in general.
To determine which parameters influence the success of a beneficial mutation, one would ideally catch beneficial mutations as they arise and follow their trajectories (whether they are successful or not) as they move through the population. Since evolution is a random process and each trajectory would represent just one specific case, one would need to observe a large number of such trajectories to illustrate general trends in how the fate of each mutation is determined. This strategy requires anticipating beneficial mutations and having a way of identifying them in the population once they arise, quickly and efficiently. In this article, we describe a method that allows us to do exactly this for a particular subset of beneficial mutations. We follow the trajectories of these mutations over the course of 1000 generations in hundreds of parallel lines and use our observations as a window into the factors that determine the fate of these spontaneously arising beneficial mutations.
Our approach relies on our previous observation that, in yeast, mutations in any one of several genes that eliminate the ability to mate provide a fitness advantage during long-term asexual propagation (Lang et al. 2009). The fitness advantage conferred by sterility is known and can be modulated by specifying the allele of GPA1, an upstream component of the mating pathway: strains carrying the ancestral (RM) allele gain a 0.6% advantage by becoming sterile, but strains carrying a derived (BY) allele common to many laboratory strains gain a 1.5% advantage. Sterility in yeast provides an ideal system for studying the evolutionary dynamics of beneficial mutations: critical parameters, such as the rate at which these mutations arise and their conferred fitness advantage, are known and can be modulated. Laboratory populations of yeast can be propagated at a range of population sizes. Furthermore, the genetic mechanisms underlying sterility are well understood. We emphasize that, since all our populations are always maintained purely asexually, sterility is not an interesting or important phenotype itself. We use it here because it allows us to observe, directly and sensitively, a common class of beneficial mutation whose dynamics we can track to gain insight into the factors influencing the fates of all beneficial mutations.
We begin by describing our method for rapidly assessing the fraction of sterile cells within a population. We next describe the details of our large-scale evolution experiment, tracking the fates of spontaneously arising sterile mutations in hundreds of evolving asexual populations at two population sizes and for two selective advantages. We show that in our populations the selective advantage of a mutation plays only a limited role in determining its ultimate fate. Instead, we find that underlying background genetic variation—not preexisting, but quickly generated by many mutations collectively in initially clonal populations—is of central importance in determining the fate of any individual mutation. In addition, we show the trajectories of individual mutations within evolving populations harbor additional information, such as the speed (and changes in the speed) of adaptation and the degree of genetic variation within the population. We also describe two other classes of repeatedly observed changes in our populations: cell aggregation and dispersed pellet morphology.
Materials and Methods
Strains
The strains used in this experiment are derived from the base strain, DBY15084, a haploid yeast strain derived from the W303 background with genotype MATa, ade2-1, CAN1, his3-11, leu2-3,112, trp1-1, URA3, bar1Δ::ADE2, hmlαΔ::LEU2. DBY15095 and DBY15092 carry a ClonNatR-marked GPA1 allele derived from RM11-1a or BY4741, respectively; this single-nucleotide change alters the amount of basal signaling through the mating pathway, thereby modulating the selective benefit conferred by sterility in these two strains (Lang et al. 2009). To allow us to use flow cytometry to detect sterility, we amplified PFUS1-yEVenus from the plasmid pNTI37 (Ingolia and Murray 2007) and integrated it at the URA3 locus of DBY15095 and DBY15092 to generate strains DBY15104 and DBY15105, respectively, using oGIL133 (5′-ACTGC ACAGA ACAAA AACCT GCAGG AAACG AAGAT AAATC CTCAC TATAG GGCGA ATTGG-3′) and oGIL134 (5′-GTGAG TTTAG TATAC ATGCA TTTAC TTATA ATACA GTTTG CAATT AACCC TCACT AAAGG-3′). Integrative transformations were performed using standard yeast procedures (Sherman et al. 1974). Sterile derivatives of DBY15095 and DBY15092 were generated, targeting the STE7 gene with the KanMX cassette (DBY15106 and DBY15107, respectively), using oGIL059 (5′-AGTTC TAAGA TTGTG TTGTC C-3′) and oGIL060 (5′-GGGTT ATTAA TCGCC TTCGG-3′). To generate a reference strain for competitive growth rate assays (DBY15108), ymCherry was amplified from pJHK044 (from John Koschwanez and Andrew Murray, Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA) and integrated at the URA3 locus of DBY15095 using oGIL270 (5′-ACTGC ACAGA ACAAA AACCT GCAGG AAACG AAGAT AAATC TCATA CACAT ACGAT TTAGG-3′) and oGIL271 (5′-GTGAG TTTAG TATAC ATGCA TTTAC TTATA ATACA GTTTG GCGGC CATCA AAATG TATGG-3′).
Long-term evolution
We propagated a total of 592 parallel cultures for 1000 generations without shaking at 30°. We varied two experimental parameters: the population size and the selective advantage of sterility. The selective advantage conferred by sterility was varied by using two ancestral strain backgrounds: DBY15104 and DBY15105. In most of this article, we refer to these strains as “RM” and “BY”, respectively; in the RM strain sterility confers a distribution of selective advantages centered around 0.6%, and in the BY strain it confers a distribution of advantages centered around 1.5% (see Results). For each population size and selective advantage we established 148 independent cultures of the ancestral strain distributed over two 96-well plates; each plate contained a unique pattern of 22 blank wells to detect contamination/cross-contamination events and to prevent plate misidentification.
All liquid handling (the dilutions, the dispensing of solutions, and the preparation for flow cytometry) was performed using the Biomek FX (Beckman Coulter, Fullerton, CA) equipped with a 16-position deck, a multichannel pod, and a Beckman Stacker Carousel, using AP96 P20 barrier for culture transfers and AP96 P250 tips for media, glycerol, and PBST transfers. The P20 tips were washed by pipetting water, ethanol, and air and then were autoclaved; tips were discarded after three uses. The big population size cultures were diluted 1:32 (4 μl into a total volume of 128 μl) every 12 hr into fresh YPD; the small population size cultures were serially diluted 1:32 × 1:32 (1:1024) every 24 hr. This propagation regime corresponds to 10 generations per day. Given a saturation density of ∼108 cells/ml, this design corresponds to an effective population size of ∼105 in the small populations and 106 in the large populations (Wahl and Gerrish 2001).
Approximately every 40 generations cultures were assayed for the presence of sterile mutants (see below) and frozen down following the addition of 50 μl of 75% glycerol to each well. During the experiment we noticed several contamination events. Immediately after starting the experiment, we observed bacterial contamination. This issue was resolved by adding ampicillin (100 μg/ml) and tetracycline (25 μg/ml) to the media. Around generation 700 we observed contamination of our media source with a standard laboratory yeast strain. This contamination was detected by observing growth in the blank wells; however, no laboratory yeast invaded the evolving populations as evidenced by plating each culture to media selective for the contaminating genotype. Twice the experiment was restarted from frozen stock (after generation 320, as a planned disruption, and after generation 820 following a possible second yeast contamination). Reanimated cultures were diluted 1:32 initially and propagated for 12 hr (5 generations) before resuming the standard propagation and dilution protocol.
Determining the frequency of sterile cells within a population
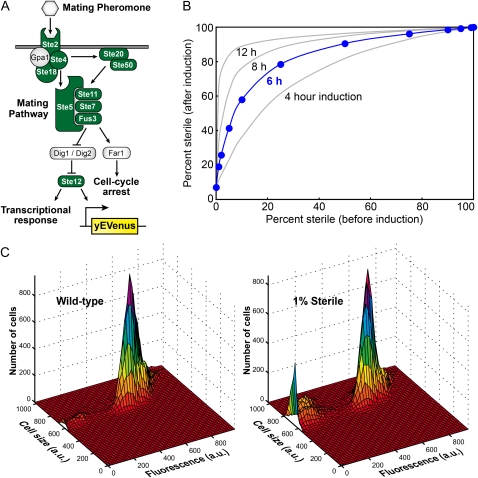
The ancestral strains, DBY15104 and DBY15105, contain a yEVenus reporter responsive to the yeast mating pheromone (αF). To detect the ratio of sterile to mating-competent cells, each plate was diluted 1:25 into a PCR plate containing 100 μl YPD + αF (10 μg/ml) per well. Plates were incubated at 30° for exactly 6 hr in a thermal cycler and then held at 4° for ∼4 hr. Plates were spun at 5000 rpm for 2 min in a Beckman Coulter centrifuge, and the medium was aspirated from the cell pellets. Cells were resuspended in cold PBST (0.5% Tween) and transferred to a 96-well plate. The ratio of yEVenus-positive to nonfluorescent cells was determined using an LSRII flow cytometer with a high-throughput sampler adaptor for 96-well plates. Since sterile cells continue to divide during the 6-hr incubation, while mating competent cells do not, the ratio of yEVenus-positive to nonfluorescent cells was converted to a frequency of steriles in the population using a standard curve (Figure 1B). To construct the standard curve, we determined the ratio of yEVenus-positive to nonfluorescent cells following αF induction for known frequencies of sterile (DBY15106 and DBY15107) strains seeded into a population of ancestral cells. The evolution experiment was begun before this method for determining the fraction of sterile cells was developed. Therefore, for data points after generation 270, the inductions were performed coincident with the dilutions, but for data points prior to generation 245, the inductions were performed on frozen cultures. In the latter case, the populations were propagated for 5 generations to acclimate the cells prior to induction; however, we noticed than many cultures failed to induce properly, leading to some spurious measurements for these early time points (supporting information, Figure S1).
Figure 1.—
Measuring the fraction of sterile cells within a population. (A) Binding of the mating pheromone (αF) is signaled through the mating pathway, ultimately resulting in a cell-cycle arrest and a transcriptional response mediated by the transcription factor Ste12. To detect mating competency, we put the fluorescent reporter yEVenus under the control of a Ste12-responsive promoter. A mutation in any one of 10 genes within the mating pathway (green) results in sterility and eliminates αF-induced expression of yEVenus. In the presence of αF, mating-competent cells arrest and induce the fluorescent reporter. Sterile cells, however, remain dark and continue dividing, thereby amplifying low-frequency steriles within a population. (B) Standard curve showing the amplification of steriles following αF inductions of 4, 6, 8, and 12 hr. For the experiments described here, we used a 6-hr induction. (C) Examples of flow cytometry profiles for cultures with no steriles (left) and with 1% sterile individuals (right) following a 6-hr αF induction.
Analyzing trajectories
We defined steriles to be “observed” if we measured a sterile frequency >0.1% in two consecutive time points. The trajectories were classified into one of the eight categories shown in Figure 3B. The initial rate of increase of sterile mutations, sup, the final rate of decrease, sdown, the generation at which sterile mutations reach 0.1%, τup, and the generation where they are forced below 0.1%, τdown, were determined by fitting the equations y = sup(t − τup) + y0 and y = sdown(τdown − t) + y0 to the upslopes and downslopes, respectively, where y is the natural logarithm of the ratio of sterile to nonsterile and t is the generation and y0 = ln(1/999), corresponding to 0.1% sterile. Fits were performed in Matlab and the points used for fitting were determined by eye. For a given trajectory, the time over which a particular sterile mutation exists at >0.1%, τtransit, is calculated as τdown − τup. The %max for each population is the highest observed percent sterile, excluding points suspected to be spurious measurements due to the freeze–thaw process (see above). For populations with multiple mutations, we recorded multiple values for the applicable parameters. All of the extracted parameters are shown in Figure S1 and Table S1.
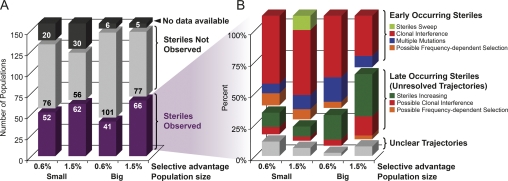
Figure 3.—
The distribution of fates of spontaneous sterile mutations. (A) The number of lines in each propagation regime in which sterile mutations were observed during the course of the experiment. Steriles were observed more frequently in small populations and when they confer a larger selective advantage. “No data available” refers to populations that were lost or that changed in a way that prohibited the detection of steriles. (B) The frequency of different types of dynamics among the lines where steriles were observed. In the larger populations, more of the trajectories remain unresolved over the 1000 generations of our experiment. This reflects the longer timescales for the spread of mutants in larger populations, but the trends in these unresolved trajectories are consistent with the rest of the data.
Fitness assays
To measure the fitness of evolved cultures from frozen stock, we compared each to a reference strain, labeled with mCherry, such that fitness is always measured against the same standard. We were careful to begin the fitness test only after both strains were growing exponentially. To this end, we thawed plates of experimental populations and mCherry-labeled reference strain (DBY15108), diluted each plate 1:32 into fresh medium, and grew them for 12 hr (5 generations) prior to mixing. The experimental and reference plates were mixed 50:50 and propagated at the appropriate population size for 30 generations. At generations 10, 20, and 30, we transferred 4 μl of saturated culture into 128 μl fresh YPD and incubated it for 3 hr. Cells were spun down and resuspended in cold PBST and the ratio of nonfluorescent (experimental) and mCherry-positive (reference) cells was determined by flow cytometry using an LSRII flow cytometer (BD Biosciences, San Jose, CA), counting 50,000 total cells for each sample. The fitness difference between the experimental and reference strain was calculated as the rate of the change in the ln ratio of experimental to reference vs. generations (Hartl 2000). To measure the fitness advantage conferred by sterility alone, we derived spontaneous steriles from the ancestral strains by plating cultures of DBY15104 and DBY15105 onto YPD + αF (10 μg/ml) and picking αF-resistant mutants from 18 independent cultures each. As described above, 50:50 mixtures of growing cultures of the mutants and the mCherry-labeled reference strain (DBY15108) were propagated in both the large and the small bottleneck regimes. Sampling and flow cytometry were performed as described above.
Fluctuation assays
Fluctuation assays (Luria and Delbrück 1943) were performed on five clones of DBY15104 and DBY15105 to determine the mutation rate to α-factor resistance as described previously (Lang and Murray 2008). Briefly, each clone was grown overnight to saturation in minimal medium supplemented with histidine, tryptophan, and uracil. Overnight cultures were diluted 1:10,000 into low-glucose minimal medium (0.1% glucose), 10 μl was dispensed into each well of a 96-well plate, and the plates were sealed with an aluminum plate seal to prevent evaporation. Cultures were grown for 36 hr at 30° without shaking, and sterile water was added to bring the volume up to 70 μl. Twenty-four cultures were pooled, diluted, and counted in triplicate using a particle counter (Beckman Coulter) to determine the number of cells per culture. The remaining 72 cultures were spot plated onto overdried YPD + αF (10 μg/ml) plates to select for mutants (9 cultures per plate) as described previously (Lang and Murray 2008). Colonies were counted after 24 hr growth at 30°. Fluctuation data were analyzed by the Ma–Sandri–Sarkar maximum-likelihood method (Sarkar et al. 1992; Foster 2006). Ninety-five percent confidence intervals were determined using Equations 24 and 25 from Rosche and Foster (2000).
Notebook
The complete laboratory notebook describing these experiments is available at http://www.genomics.princeton.edu/glang/notebooks.htm.
Results and Discussion
We showed previously that mutations within the yeast mating pathway provide a growth rate advantage and that this benefit can be modulated such that sterility results in either a 0.6% (RM) or a 1.5% (BY) fitness advantage (Lang et al. 2009). Using strains with a fluorescent reporter of mating ability, we initiated 148 parallel experimental populations for each of four propagation regimes: small populations (Ne = 105) in which sterility provides a 0.6% fitness advantage (RMS populations), small populations where sterility provides a 1.5% advantage (BYS), big populations (Ne = 106) where sterility provides a 0.6% advantage (RMB), and big populations where sterility provides a 1.5% advantage (BYB). Each initially clonal population was propagated in a static environment by daily (or twice-daily) serial transfer for 1000 generations and assayed for the percentage of sterile cells in each population approximately every 40 generations.
Of our 592 total experimental lines, a total of 61 were lost over the course of the experiment, primarily due to the evolution of phenotypes that interfered with our ability to assay the frequency of steriles using flow cytometry. We describe some of these in more detail below. In the remaining 531 populations, we observed steriles at detectable frequencies in 221 populations across the four regimes. A complete description of all 221 populations is provided in Figure S1.
The fates of beneficial mutations
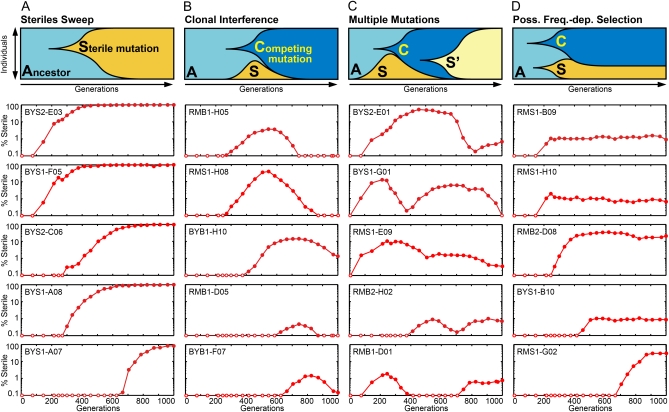
Once steriles were observed, they experienced four general fates. The simplest case is a selective sweep: a spontaneous sterile mutation arises and increases in frequency until it fixes (Figure 2A). Selective sweeps were observed in only 8 of 221 cases, only at the small population size of the strain in which steriles provide a 1.5% advantage. More commonly, sterile alleles rose to some frequency, but were outcompeted by a more-fit lineage before they were able to fix (clonal interference, Figure 2B). More complicated trajectories were also observed, in which sterile strains rise, are subjected to clonal interference, and then increase in frequency again (Figure 2C). This could reflect a second beneficial mutation occurring in a declining sterile population or a second sterile mutation occurring in the background of the competing mutation (as shown). Evidence for these three types of trajectories has been observed in a variety of other experimental studies, such as the recent work by Miller et al. (2011). We were surprised to also observe a fourth type of trajectory where sterile strains rise to some frequency and remain there for hundreds of generations, suggesting the action of frequency-dependent selection (Figure 2D). Of 50 putative examples of this type of trajectory, 38 were deemed spurious because they gave a false signal due to changes in the response to αF. The 12 examples reported here appear to be genuine examples of frequency-dependent selection. We are currently exploring this phenomenon; a more complete description of these populations will be presented elsewhere.
Figure 2.—
Spontaneous sterile mutations experienced one of four general fates. The top row illustrates a hypothetical scenario that could produce each of the four types of dynamics. Below each illustration are five representative examples of each dynamic. For each experimental population, the frequency of sterile mutants is shown as a function of time and the populations are identified by their systematic names (Figure S1). Note the logarithmic y-axis. (A) The simplest case is a selective sweep. (B) More commonly, sterile mutations are outcompeted by more fit lineages, a process known as clonal interference. (C) In some cases we observed the reemergence of steriles after clonal interference. This could occur either by a second sterile mutation (as shown in the illustration at the top) or by an additional beneficial mutation arising in the original sterile lineage. (D) Long-term maintenance of sterile mutations at a given frequency could indicate the action of frequency-dependent selection, where the difference in fitness between the sterile and nonsterile subpopulations is a function of the frequency of the subpopulations. This phenomenon is currently being explored and will be reported elsewhere.
Since each trajectory represents only one possible outcome, we looked at the frequency at which each type of outcome was observed across our many independent populations to illustrate general trends in how the fate of beneficial mutations depends on the population size and on the selective advantage that they provide. This distribution of outcomes is illustrated in Figure 3. To rule out the possibility that our observations of the outcomes and dynamics of sterile mutations in BY and RM lines are due to difference in mutation rates in the two strains used in this experiment, we measured the mutation rate to sterility in the ancestral strain backgrounds (Table 1), using the fluctuation method of Luria and Delbrück. We found no difference in the mutation rates to sterility in the strain backgrounds producing 0.6% and 1.5% mutations: 2.37 × 10−6 and 2.27 × 10−6, respectively. This confirms that the difference in the observed dynamics of 0.6%- and 1.5%-effect sterile mutations can be attributed to the differences in the fitness effect and not in the supply of mutations.
Table 1.
Mutation rate to sterility (αFR) in ancestral strains
| Straina | Clone | Mutation rate to αFR (×10−6 per generation) |
| DBY15104 (sste = 0.6%) | A | 2.68 (1.89–3.58, 95% confidence interval) |
| B | 2.50 (1.75–3.35) | |
| C | 2.17 (1.49–2.94) | |
| D | 2.19 (1.50–2.98) | |
| E | 2.30 (1.59–3.10) | |
| A–E combined | 2.37 (2.03–2.73) | |
| Average ± SD | 2.37 ± 0.22 | |
| DBY15105 (sste = 1.5%) | A | 2.85 (2.00–3.80) |
| B | 2.51 (1.74–3.37) | |
| C | 1.88 (1.26–2.59) | |
| D | 2.20 (1.53–2.96) | |
| E | 1.96 (1.31–2.71) | |
| A–E combined | 2.27 (1.94–2.62) | |
| Average ± SD | 2.28 ± 0.40 | |
| yGIL104 (Lang and Murray 2008) | 5.86 (5.46–6.28) | |
| DBY15084 (Lang et al. 2009) | 2.79 (2.24–3.38) |
The strains used in this experiment (DBY15104 and DBY15105) as well as the strain DBY15084 were derived from yGIL104. The silent mating cassette (HMLα) is present only in yGIL104. All of these strains can become resistant to αF by mutation; however, yGIL104 can also become resistant to αF by mating-type switching or by expression of the silent mating cassette. The rate of αF resistance in this strain is 5.86 × 10−6, suggesting that in HMLα strains, the rate of production of αF-resistant strains, by way of mating-type switching or by expression of the silent mating cassette, is ∼3.5 × 10−6.
Adaptation is not mutation limited
If adaptation were mutation limited, sterile mutations would occur at random times and increase in frequency at a rate given by their fitness advantage until they sweep to fixation (Ewens 2004). We would see sterile mutations ∼10 times more often in our big populations (although this is a slight overestimate, since sterile mutants need to rise to larger absolute numbers to be at a detectable 0.1% frequency in big populations than in small). We would also see them more than twice as often in BY populations than in RM, since their probability of surviving genetic drift to reach detectable frequencies is approximately proportional to their selective advantage. Further, they would spread more quickly in our BY lines where they provide a larger selective advantage.
None of these null expectations held true for our populations. Rather, selective sweeps were rare, and clonal interference was a far more likely fate for any particular mutation. Sterile mutations were clearly not observed more often in big populations; if anything, they were observed more often as a fraction of the total number of lines in small populations, although this difference is minor and not statistically significant (P = 0.13, chi-square test). While we observed steriles somewhat more frequently in BY than in RM populations, the difference is much less than expected given that 1.5%-effect mutations are more than twice as likely to survive drift as 0.6%-effect mutations.
All of these observations, particularly the extensive observations of clonal interference, suggest that our populations are in the regime where many beneficial mutations occur and spread simultaneously, as in many earlier experimental studies (De Visser et al. 1999; Miralles et al. 1999; De Visser and Rozen 2006; Desai et al. 2007; Gresham et al. 2008; Kao and Sherlock 2008; Miller et al. 2011). In this regime, mutation rapidly generates variation in fitness within a population (Desai and Fisher 2007; Desai et al. 2007; Rouzine et al. 2008). Where within this background of variation a given mutation (and future competing mutations) falls can slow down or speed up the spread of a given mutation and determine its ultimate fate (Desai and Fisher 2007; Park and Krug 2007; Fogle et al. 2008; Park et al. 2010). To investigate these effects further, we conducted a more detailed analysis of the dynamics of each observed sterile mutation.
Parameterization of mutational trajectories
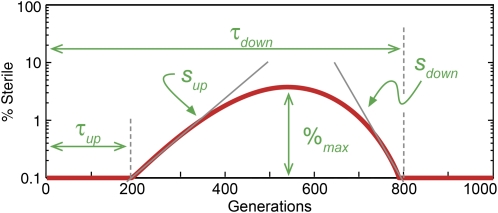
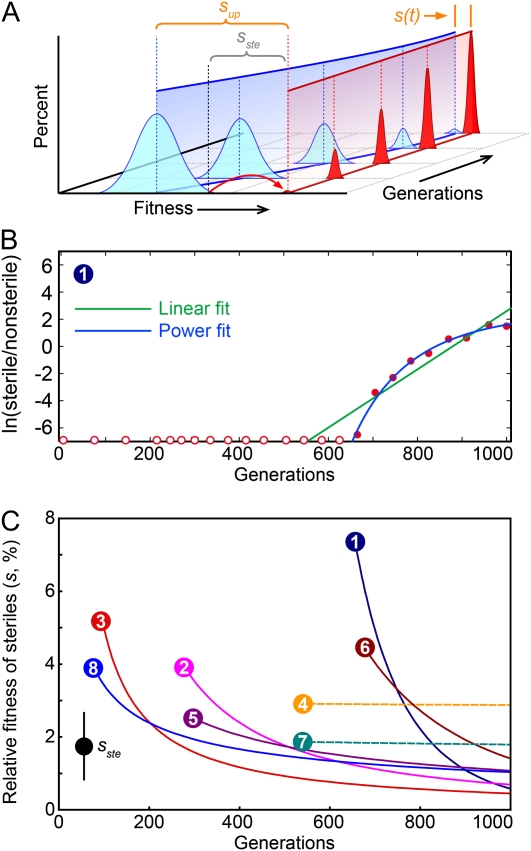
In addition to the distribution of fates of sterile mutations across our four propagation regimes, the specific trajectory of each mutation contains quantitative information about the dynamics of adaptation in each individual population. Together, this illustrates the role of genetic variation on the emergence and fate of beneficial mutations. To access this information, we described the trajectory of each observed sterile mutation by a set of parameters (Figure 4). We described the exponential spread of steriles through a population with two parameters: τup, the time at which steriles are initially observed upon reaching a frequency of 0.1%; and sup, the initial exponential rate of increase (the “upslope”). Similarly, we parameterized the decline of steriles in a population by the time at which the sterile fell below detectable frequencies, τdown, and by the “downslope,” sdown. We denote the maximum frequency ever reached by the sterile mutation by %max (note that %max is an underestimate if the sterile frequency is still increasing at the end of our experiment). For populations with more complex dynamics involving multiple upslopes and/or downslopes, we compute the multiple values of these parameters and distinguish between them with subscripts.
Figure 4.—
A schematic of a clonal interference trajectory illustrating the parameters measured for each trajectory. The initial spread of steriles is characterized by the time at which steriles initially reach 0.1% (τup) and the initial exponential (linear in log space) rate of increase (sup). Similarly, the purging of steriles is characterized by sdown and τdown. The maximum height of the trajectory is %max. Not all parameters were extracted from each trajectory, and for some trajectories, multiple values for a given parameter were obtained where possible and are denoted by subscripts. The full list of extracted parameters is available in Figure S1 and Table S1.
The emergence of sterile mutations
We begin by considering the initial rate of spread of sterile mutations (the upslope, sup). The null expectation is that the distribution of sup will reflect the fitness advantage provided by sterility. This is expected if sterile mutations occur in populations that lack variation in fitness. In the presence of other genetic variation, however, the initial rate at which the sterile mutation accumulates, sup, measures the fitness of the individual containing this sterile mutation relative to the mean fitness in the population at that time (Figure 5A). This tells us where within the background of genetic variation the mutation occurred.
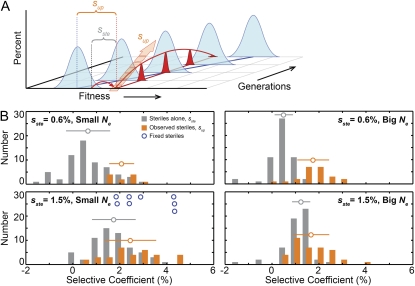
Figure 5.—
The distribution of sup and the role of genetic variation. (A) A three-dimensional representation of a sterile trajectory (in this case, involving clonal interference) showing that sup is a measure of the fitness of the sterile lineage relative to the population mean fitness at the time the sterile mutation arose. That is, sup is the sum of the fitness of the background in which the sterile mutation occurred and of the fitness advantage provided by the sterile mutation itself. (B) The distribution of fitness effects of the sterile mutation alone in the ancestral background (sste, gray bars) centers on 0.6% (top) and 1.5% (bottom) in the RM and BY lines, respectively. In all four regimes, however, the initial rate of spread of sterile mutations (sup, orange bars) is faster than can be explained by the effect of the sterile mutation alone, sup > sste (P = 2.5 × 10−7, 2.7 × 10−3, 4.4 × 10−9, and 2.6 × 10−3, for 0.6% small Ne, 1.5% small Ne, 0.6% big Ne, and 1.5% big Ne, respectively, two-tailed t-test, unequal variance). Surprisingly, the distribution of sup is independent of the fitness advantage of the sterile mutation itself (P = 0.66 and P = 0.13 for the hypothesis that sup depends on fitness for big and small Ne, respectively, two-tailed t-test, unequal variance). The eight populations in which steriles swept to fixation are indicated by open blue circles; three of these have sup > 4%. The open circles and horizontal bars indicate the means and standard deviations of the distributions.
To measure the fitness effect of sterile mutations alone, we isolated spontaneous steriles derived from the ancestral strains and measured their fitness under the experimental conditions (sste, gray bars in Figure 5B). The distributions of sste show some variation centered on 0.6% and 1.5%, respectively. This variation is substantially larger than the errors in our fitness measurements and presumably reflects biological variation due to the fact that not all sterile mutations have precisely the same effect.
In Figure 5B, we compare our null expectation (gray bars) to the actual distribution of upslopes in our four propagation regimes (orange bars). We see that the null expectation is clearly incorrect: in all four regimes steriles initially accumulated faster than would be expected given the fitness advantage provided by a sterile mutation alone, indicating that only sterile mutations that occur in favorable genetic backgrounds reach even modest frequencies in the population. Strikingly, the average sup in all four regimes is similar (∼2%), indicating that the initial rate at which steriles accumulate in the population does not depend on whether the sterile mutations themselves provide a 1.5% or a 0.6% fitness advantage.
Genetic variation and the emergence of beneficial mutations
This observation points to the underlying variation as the determinant of sup: we infer that those sterile mutations that survive and accumulate occurred in cells already more fit than most of the population as the result of the accumulating background genetic variation. Mutations conferring sterility are only a small subset of possible beneficial mutations, so this genetic variation is similar regardless of the fitness advantage provided by the sterile mutation itself. In all populations, genetic variation is quickly created and mutants with an ∼2% fitness advantage initially tend to be among the most fit in the population and thus increase in frequency. We observe steriles when they combine with other mutations to reach this threshold. Therefore, to be observed, a sterile mutation conferring a 0.6% fitness advantage must occur in a fitter background than one conferring a 1.5% advantage: either farther out in the tail of the fitness distribution or at times when genetic variation is higher (occasionally a sterile mutation may occur first, and then other genetic variation develops around it; this is not inconsistent with our discussion, but it is presumably rare because steriles represent a small fraction of possible beneficial mutations, and there are many populations where we never observe them at all). Thus while the fitness advantage provided by the sterile mutation influences the probability it will reach a high enough frequency to be observed, the fitness advantage of the clones containing the sterile mutations and hence the subsequent dynamics are almost independent of the advantage provided by the sterile mutation itself.
Note that in a few cases a sterile mutation will occur in a very fit individual in the background where it provides a 1.5% advantage and thus become part of a clone that is more fit than would be attainable if the sterile provided only a 0.6% advantage. This means that the maximum rates of increase of steriles across all 531 lines were observed in populations where steriles provide a 1.5% advantage, and correspondingly steriles fix only in these populations; we discuss this further below. This happens rarely, however, and in the typical population the dynamics are indeed independent of the fitness advantage provided by the sterile mutation itself.
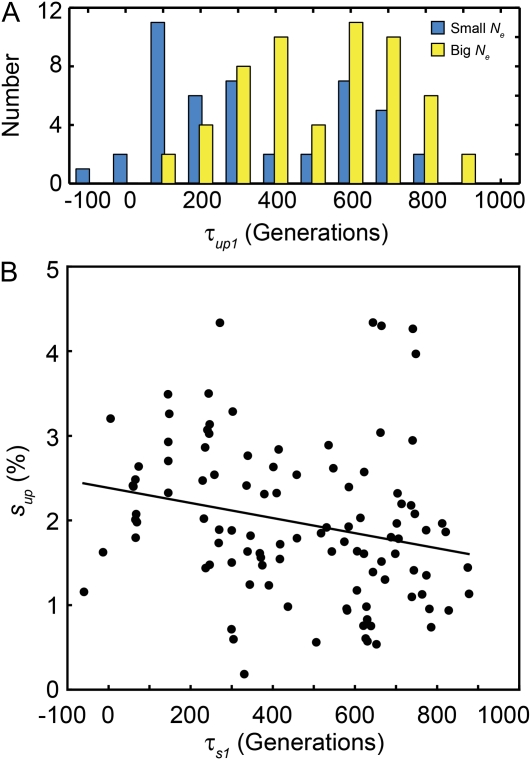
Presumably underlying genetic variation takes some time to develop and may fluctuate over time. The initial upslope of steriles, however, is only very weakly correlated with the time in which that sterile mutation arose, and in fact the upslopes for steriles that arise early on in the experiment are if anything slightly larger than those that arise late (Figure 6). This indicates that the underlying genetic variation that drives our observations about the distributions of upslopes is generated within at most ∼100 generations and does not experience significant systematic changes over the duration of our 1000-generation experiment.
Figure 6.—
The waiting time to the first observation of sterile mutations, τup1. (A) The distribution of τup1 shows that sterile mutations were initially observed earlier in the small Ne populations (blue bars) than in big Ne populations (yellow bars). (B) Late-occurring steriles spread slightly slower than early-occurring steriles (P = 0.0016, Spearman's rank correlation). The black line is the best-fit linear regression (Pearson's) to the data.
The most successful mutations are the luckiest
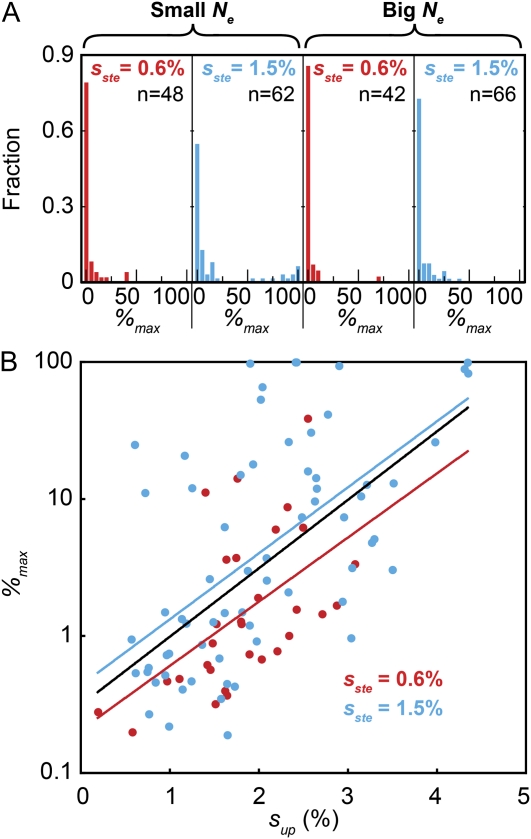
We have thus far drawn a clear distinction between “successful” sterile mutations that experience selective sweeps and those that fall victim to clonal interference. Even among those sterile mutations destined for elimination by clonal interference, there is substantial variation in their overall success, as measured by the maximum frequency they attain in the population, %max (Figure 7A). As might be expected, those mutations that reach the highest frequencies in our populations tend to be those that had the highest initial upslopes—that is, sup and %max are positively correlated (Figure 7B). This observation makes sense in terms of the role of underlying genetic variation: those mutations that have the largest sup are those that occurred in the best genetic backgrounds, and thus they were able to reach higher frequencies before the mean fitness of the rest of the population outstripped them. In other words, the most successful mutations are those that were lucky to occur in the best genetic backgrounds; this is consistent with results from earlier work (Miller et al. 2011).
Figure 7.—
The maximum frequency attained by sterile mutations, %max. (A) The distribution of %max across the four regimes shows that sterile trajectories attained greater frequency in smaller populations and when sterile mutations confer a larger selective advantage. (B) There is a strong positive correlation between sup and %max (P < 10−5, Spearman's rank correlation on the combined data). The black line is the best-fit linear regression (Pearson's) to the data. Separating the data from RM and BY lines still produces a significant positive correlation (P = 0.0008 and P < 10−5 for sste = 0.6% and 1.5%, respectively, Spearman's rank correlation). However, the best-fit line to the sste = 1.5% data is shifted to a higher %max relative to the best-fit line to the sste = 0.6% data. Sterile mutations with a higher fitness advantage, therefore, attain a higher %max.
Larger-effect mutations can attain higher frequency
Although the average sup is similar for the four propagation regimes, nearly one-fifth (12/62) of the 1.5%-effect mutations in small Ne populations have %max values >50%, while in the other three propagation regimes combined only 1 of 156 populations reached at least 50% sterile (Figure 7A). How does one reconcile this discrepancy? Although the average upslopes are similar in all four regimes, the variation in sup is greater in the BY populations. Since %max is correlated to sup (Figure 7B), differences in upslopes can contribute to the %max in the BY populations. Indeed, three of the eight BYS populations that fixed steriles have an sup > 4% (Figure 4D). The correlation between sup and %max, however, accounts for only 40% of the variation in %max, indicating that other factors contribute to suppressing the spread of 0.6%-effect mutations. To generate a sterile clone with fitness 2% greater than the population mean, a 1.5%-effect sterile mutation could occur in a population with less variation in fitness than is needed for a 0.6%-effect sterile mutation. Since the rate of adaptation is equal to the variance in fitness within a population (Fisher 1930), increased variation in RM populations will more rapidly slow the spread of the sterile lineage. We show below that it is indeed the case that 0.6%-effect sterile mutations are observed to emerge from the population at times when the genetic variation is high. This bias explains why smaller-effect mutations are more susceptible to competing beneficial mutations and are less likely to reach high frequency.
Genetic variation and the dynamics of selective sweeps
We observed selective sweeps in eight populations from the small population size lines where steriles confer a 1.5% advantage. We expect that genetic variation in fitness at the time the sterile mutation arose should affect the dynamics of the subsequent selective sweeps (Figure 8A). Specifically, if all cells in the population were equally fit at the time of the sterile mutation, the fitness difference between the steriles and nonsteriles will be approximately constant throughout the duration of the sweep. This means that the rate of change of the log ratio of steriles to nonsteriles should be constant over the course of the sweep. In contrast, if genetic variation is present, the difference in fitness between the steriles and nonsteriles will decrease during the sweep as the lower-fitness nonsteriles are purged from the population (note that lower-fitness steriles will also be purged, but compared to the nonsteriles, fitness variation is likely to be small in the sterile subpopulation due to its recent bottleneck and corresponding small initial population size).
Figure 8.—
Schematic of a selective sweep. (A) A schematic of a selective sweep showing that, unlike with clonal interference, the sterile mutation initially occurs in a highly fit individual and is never outrun by another lineage. If, at the time the sterile mutation arose, there was variation in fitness within the population, the relative fitness of the sterile lineage will decrease during the sweep as the mean fitness of the nonsterile subpopulation increases. By the end of the sweep, the sterile lineage will be competing against a population similar in fitness to the background in which the sterile mutation arose. (B) An example of a selective sweep, shown as the change in the log ratio of steriles to nonsteriles over time fit to a linear model (corresponding to a constant relative fitness of the sterile lineage) and a power law model (corresponding to a decreasing relative fitness of the sterile lineage). Similar plots for the other seven sweep populations are shown in Figure S2. (C) The change in the relative fitness of the sterile lineage relative to the population mean fitness can be evaluated at any point by calculating the derivative of the best power-law fit to the data. The initially fast increase slows to a value similar to the fitness provided by the sterile mutation alone, sste, by the end of the sweep. Populations 1–8 are BYS1-A07, -A08, and -F05 and BYS2-C03, -C06, -D06, -D07, and -E03, respectively.
To distinguish between these two possibilities, we fit the trajectories of the selective sweeps to models assuming either uniform or decreasing fitness of the steriles relative to the rest of the population (Figure 8B and Figure S2). For six of the eight populations, the fitness advantage of the sterile mutants declines over time, until near the end of the sweep the steriles are spreading at a rate consistent with them competing against a nonsterile subpopulation similar in fitness to the background in which the sterile mutation arose (Figure 8C, solid lines). This is consistent with the sterile mutation arising in a very favorable genetic background and initially increasing at a rate much faster than the sterile mutation alone would allow. For the other two populations the fitness of the steriles is constant, consistent with the sterile mutation alone arising in a population without extensive genetic variation, for example on the heels of a strong selective sweep (Figure 8C, dashed lines). Of the 531 populations analyzed in this study, these are the only two consistent with simple models of adaptation where a beneficial mutation arises and sweeps through an otherwise isogenic population.
A new take on clonal interference
The rate at which steriles spread typically slows down over time, as the mean fitness of the rest of the population increases due to selection on existing variation or new mutations. This leads to either a slowing down of a selective sweep or clonal interference. Clonal interference occurs when the mean fitness of the population oustrips that of the sterile clone, and the rate of spread of the steriles becomes negative. This can occur either because the sterile mutation was never the most-fit individual in the population (i.e., existing variation) or because subsequent mutations outstripped it (i.e., new mutations). In the case of a selective sweep, the sterile mutation generally occurs in a very favorable genetic background and thus increases in frequency much faster than the sterile mutation alone would allow. During the sweep, the nonsterile subpopulation increases in fitness relative to the sterile subpopulation, but never outstrips it. The rate of increase of the steriles therefore declines over time (Figure 8C).
Our results show that although selective sweeps and clonal interference are strikingly different outcomes from the point of view of a particular mutation, they are both special cases of how individual new mutations interact with the existing variation in the population. The difference between sweeps and clonal interference is, in this view, largely determined by where within the distribution of underlying variation the cell that acquires the sterile mutation lies. A sweep will be likely only if the sterile mutation occurs in a very fit cell; if the sterile mutation occurs in a less fit cell, a sweep is unlikely and clonal interference is the most probable outcome. This reinforces the importance of underlying genetic variation and stands in contrast to the traditional viewpoint of clonal interference as a phenomenon caused mainly by later mutations (Gerrish and Lenski 1998; Wilke 2004).
The tempo of adaptation
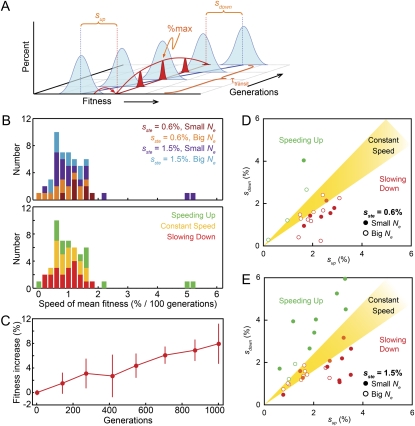
Many previous experiments in a variety of microbes have measured the rate of adaptation during long-term evolution experiments. These methods have typically involved direct measurements of the mean fitness of the populations over time. The trajectories of individual sterile mutations give us an alternative way to measure the speed of adaptation. As illustrated in Figure 9A, the parameters sup, sdown, and %max provide a measure of the fitness of the sterile subpopulation relative to the population mean fitness at specified times. Specifically, sup records how far ahead of the population mean fitness the sterile subpopulation was at the time at which steriles first reached 0.1% (τup), sdown records how far behind the population mean fitness the sterile subpopulation was when it returned to 0.1% (τdown), and %max is reached at a time τmax when the fitness of the sterile subpopulation is equal to the population mean fitness. It follows that the average speed of mean fitness increase over the time in which steriles were detectable is (sup + sdown)/τtransit, where τtransit = τdown – τup.
Figure 9.—
Rate of mean fitness increase. (A) A schematic of a clonal interference trajectory relating the parameters sup, sdown, and %max in terms of the fitness of the sterile subpopulation relative to the mean fitness of the population. The average speed of mean fitness increase is (sup + sdown)/τtransit, where τtransit = τdown – τup. If the speed of mean fitness increase is constant, the trajectory will be symmetrical with sup = sdown and %max occurring equidistant between τup and τdown. Asymmetry indicates either the slowing down (sdown < sup) or speeding up (sdown > sup) of the speed of mean fitness increase. (B) The average rate of mean fitness increase during the transit time of the sterile lineage, as inferred from the 51 high-quality clonal interference trajectories, color coded by propagation regime (top) and acceleration of mean fitness increase (bottom). (C) The rate of adaptation for 12 of the 51 populations for which standard (competitive fitness assay) measurements of mean fitness over time were available. This independent method of calculating the speed of mean fitness increase is consistent with the estimate of 1%/100 generations as estimated from the trajectories. (D) Relationship between sup and sdown in populations in which we observed 0.6%-effect sterile mutations; note the excess of trajectories in which the speed of mean fitness increase is slowing down. (E) Relationship between sup and sdown in populations in which we observe 1.5%-effect sterile mutations; these are not biased in terms of changes in mean fitness increase.
For the 51 trajectories where we have reliable measurements for both sup and sdown, we used this method to calculate the speed of mean fitness increase. We find that in all four regimes, mean fitness increases by ∼1% per 100 generations (Figure 9B). We compared these results to direct measurements of the changes in mean fitness of a subset of our populations every 140 generations (including 12 of the 51 populations shown in Figure 9B), as measured with standard competitive fitness assays (see Materials and Methods). The rate of adaptation as measured by these direct growth rate assays is consistent with our estimates from the trajectories themselves (Figure 9C).
Using our results, we can also investigate changes in the rate of adaptation during the time that a sterile mutation was present in our populations. If the speed of mean fitness increase is constant during this time, we expect that a trajectory in which sterile mutations underwent clonal interference should be symmetrical, with sup = sdown and with %max occurring at the halfway point in the trajectory (as shown in Figure 9A). On the other hand, differences in sup and sdown indicate that the speed of mean fitness increase has not been constant over the duration of the trajectory: sdown > sup implies that mean fitness increase is speeding up, and sdown < sup implies that it is slowing down.
In Figure 9, D and E, we show the relationship between sup and sdown for each trajectory. We see that there is substantial variation in whether the rate of adaptation is speeding up or slowing down between individual populations. Changes in the speed of mean fitness increase indicate changes in the variation of fitness within the population (Fisher 1930). Not only is genetic variation important in determining the fate of individual beneficial mutations, but also this variation itself fluctuates according to the stochastic nature of mutational events.
What is the significance of these fluctuations? The fate of individual beneficial mutations depends on whether they happen to occur at times of high or low variability. Small-effect mutations require more underlying genetic variation to produce a clone with the same fitness as a large-effect mutation. This results in an observation bias in that 0.6%-effect mutations tend to be observed at times of expanded genetic variation (i.e., when the mean fitness increase is fast and will likely slow down in the future, Figure 9D) relative to 1.5%-effect mutations. As described above, this bias can explain why 1.5%-effect sterile mutations attain a higher %max compared to 0.6%-effect sterile mutations, despite similar distributions of sup.
It is a useful analogy to consider this process of adaptation as a traveling wave of fitness, where transient increases in the width of the traveling wave result in a concomitant increase in the speed of mean fitness increase, which in turn reduces the variation in fitness in the population. The stochastic nature of mutation, and the distribution of fitness effects of these mutations, regulates the breathing and faltering progression of the traveling wave.
Aggregates and dispersive cell pellets
During the course of the 1000-generation evolution, we noted several phenotypic changes other than sterility that arose spontaneously in our populations. These included aggregations of cells in a few of our lines and dispersive cell pellets in others.
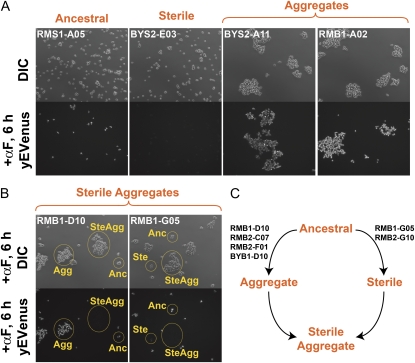
Cell aggregates in a few evolving lines were first observed during flow cytometry and were verified by microscopy. Figure 10A shows examples of cell morphologies in these aggregating populations. We observed aggregation in 52 populations across the four regimes. Note that many of these populations were excluded from the analysis of the dynamics of sterile mutations since aggregates that arose early in the evolution prohibited quantitative assessment of sterile frequencies by flow cytometry (these populations fall into the “no data available” category in Figure 3). Nevertheless, we did observe sterile aggregates in 6 populations. There are two ways in which this could occur: sterility arose first, followed by aggregation, or vice versa. Figure 10B shows an example of each. Both the top images (light microscopy) and the bottom images (fluorescence) were taken on the same field of cells following αF induction. On the basis of the presence of three cell types (ancestral, aggregates, and sterile aggregates) in population RMB1-D10 we can infer that the mutation conferring aggregation occurred first and that the sterile mutation arose in this background. Conversely, for population RMB1-G05, the presence of steriles and sterile aggregates (but not nonsterile aggregates) indicates that the sterile mutation occurred first and that the aggregate mutation occurred in the sterile background. For 4 of the 6 populations where sterile aggregates were observed, the aggregate mutation occurred first (Figure 8C).
Figure 10.—
Selection for cell aggregation. (A) Examples of cell morphologies in four populations after 1000 generations. The top panels correspond to light images of each population and the bottom panels are fluorescence images of the same populations following 6 hr of αF induction. Population RMS1-A05 retained the ancestral phenotype with respect to sterility and aggregation, BYS2-E03 fixed a sterile mutation, and BYS2-A11 and RMB1-A02 evolved aggregation. Aggregation was observed in 52 populations across the four regimes (note that many of these populations were excluded from the analysis of the dynamics of sterile mutations and fall into the “no data available” category in Figure 3 since aggregation interferes with the quantification of sterile frequencies). Nevertheless, we observed sterile aggregates in 6 populations. (B) Two examples of sterile aggregates. Both the top images (light microscopy) and the bottom images (fluorescence) were taken on the same field of cells following αF induction. There are two ways in which sterile aggregates can arise: sterility first or aggregation first. On the basis of the presence of three cell types (ancestral, aggregates, and sterile aggregates) in population RMB1-D10 we can infer that the mutation conferring aggregation occurred first and that the sterile mutation arose in this background. Conversely, for population RMB1-G05, the presence of steriles and sterile aggregates (but not nonsterile aggregates) indicates that the sterile mutation occurred first and that the aggregate mutation occurred in the sterile background. (C) For 4 of the 6 populations where sterile aggregates were observed, the aggregate mutation occurred first.
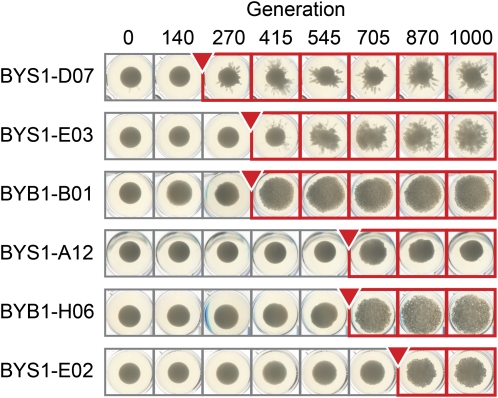
During the evolution experiment, the 96-well plates were not shaken (although they were mixed at each dilution), making it possible for spatial structuring to emerge (Figure 11). For two of the eight 96-well plates (BYS1 and BYB1) we imaged the cell pellets prior to mixing and dilution, approximately every 140 generations (Figure S3). In general, pellet size increased during the 1000-generation experiment. Among these 144 populations we observed 17 with dispersed pellet morphologies.
Figure 11.—
Selection for dispersed cell pellet morphologies. Six examples of heritable changes in cell pellet morphology from plates BYS1 and BYB1 are shown. Imaged cell pellets from all 144 populations on these two plates are shown in Figure S3.
Both aggregation and dispersed pellet morphologies are beneficial in our propagation regime. In the small Ne populations, when these phenotypes were observed, they swept to fixation (Figure S3). The same is true for aggregates in the big Ne populations. Abnormal pellet morphologies, however, frequently arise and are outcompeted at the big population size. By contrast, the sterile mutations fix only in the small Ne populations and are almost always outcompeted at either population size (there is, of course, an observational bias here, in that steriles can be detected at much lower frequencies than aggregates of dispersed pellet morphologies). We identified several examples of the rise of these other alternative phenotypes and the concomitant loss of steriles by clonal interference (Figure S4). These observations suggest that dispersed pellet morphologies are more fit than steriles and that aggregates are more fit than either of those in our propagation regime.
The utility of mutation-tracking experiments
Most long-term evolution experiments focus retrospectively on a small number of successful clones. These studies are informative in determining which mutations confer a selective advantage, whether the order of mutations matters, and how many pathways to adaptation exist for a given condition. Some aspects of the dynamics of evolution can be gleaned from these studies. Measuring fitness at several time points reveals the average rate of adaptation, and measuring the fitness of many clones at a given time point provides a snapshot of genetic variation within the population. Many of the details of the dynamics, however, are not accessible to this type of analysis.
The mutation-tracking experiment presented here provides an alternative strategy to studying evolutionary dynamics that, in many ways, is the opposite of the retrospective approach: rather than analyze the successful clones emerging from the population, we track a subset of beneficial mutations whether they are successful or not. We contend that mutation-tracking experiments, especially when combined with high-throughput maintenance of a large number of parallel cultures, provide a way of addressing aspects of the evolutionary process inaccessible to previous methods.
Although mutation-tracking experiments can, in principle, be done with any mutation (or class of mutation) for which a reporter system can be devised, sterility in yeast is in many ways an ideal system. The mutation rate to sterility is high enough that we see steriles regularly, but not so high as to often result in multiple steriles arising and spreading simultaneously. The fitness advantage of steriles is known and can be modulated. The phenotype can be selected for in the presence of αF; this allows for mutation rate estimation by fluctuation assays, for the isolation of spontaneous sterile mutations in the ancestral background, and provides a means for amplifying these mutations in culture. The 6-hr αF induction (necessary to induce the fluorescent reporter in the nonsterile cells) has the ancillary benefit of extending the dynamic range of detection and allowing observation of trajectories at low frequency, when many of the important dynamics are occurring. Increasing the induction time beyond 6 hr can further extend this dynamic range (Figure 1B). We commented in several places above regarding observation biases in the current experiment, namely that we observe steriles only when they fall in favorable backgrounds. Extending the dynamic range could reveal very low-frequency steriles in the population and perhaps reveal more details of the dynamics of adaptation. Continued careful measurement of these trajectories, combined with a sensible interpretation of their meaning, has the potential to greatly improve our understanding of the dynamics of adaptation over short timescales.
In this article, we have used the details of the trajectories of sterile mutants to show how genetic variation plays an essential role in the evolutionary dynamics of each mutation. A variety of recent experimental work has shown that in large populations beneficial mutations can be very common (De Visser et al. 1999; Miralles et al. 1999; Joseph and Hall 2004; De Visser and Rozen 2006; Desai et al. 2007; Gresham et al. 2008; Kao and Sherlock 2008; Miller et al. 2011), which has led to a focus on the dynamics of individual new mutations and the competition between them (Gerrish and Lenski 1998; Wilke 2004; Desai and Fisher 2007). Our results show that the collective formation of genetic variation due to all these new mutations influences the fate of any one of them. Although adaptation by new mutations vs. adaptation from standing genetic variation are sometimes thought of as distinct mechanisms (Barrett and Schluter 2008), we have found that even in initially clonal populations, new mutations can generate variation rapidly enough (even while selection acts) to strongly influence the dynamics of adaptation. Together, our observations paint a picture of the importance of underlying variation in determining the fate of individual beneficial mutations, whose dynamics are determined by how well they piggyback on existing variation and how quickly they are outcompeted by it.
Acknowledgments
We thank Ted Cox, Mark Rose, Aleksandra Walczak, Chris Marx, Andrew Murray, Bodo Stern, and members of the Botstein and Desai labs for useful conversations and comments on the manuscript. We thank J. Buckles and D. Storton for assistance with robotic liquid handling and C. DeCoste for assistance with flow cytometry. This work was supported by a National Institute of General Medical Sciences Centers of Excellence grant GM071508 (to D.B.), an individual National Institutes of Health grant GM046406 (to D.B.), and a 21st Century Science Initiative Grant from the James S. McDonnell Foundation (to M.M.D.).
Literature Cited
- Barrett R. D. H., Schluter D., 2008. Adaptation from standing genetic variation. Trends Ecol. Evol. 23: 38–44 [DOI] [PubMed] [Google Scholar]
- Barrick J. E., Lenski R. E., 2009. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 74: 119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., et al. , 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247 [DOI] [PubMed] [Google Scholar]
- Betancourt A. J., 2009. Genomewide patterns of substitution in adaptively evolving populations of the RNA bacteriophage MS2. Genetics 181: 1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollback J. P., Huelsenbeck J. P., 2007. Clonal interference is alleviated by high mutation rates in large populations. Mol. Biol. Evol. 24: 1397–1406 [DOI] [PubMed] [Google Scholar]
- Buckling A., MacLean R. C., Brockhurst M. A., Colegrave N., 2009. The Beagle in a bottle. Nature 457: 824–829 [DOI] [PubMed] [Google Scholar]
- De Visser J., Zeyl C. W., Gerrish P. J., Blanchard J. L., Lenski R. E., 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283: 404–406 [DOI] [PubMed] [Google Scholar]
- De Visser J. A. G. M., Rozen D. E., 2006. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics 172: 2093–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., 2007. Beneficial mutation-selection balance and the effect of linkage on positive selection. Genetics 176: 1759–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., Murray A. W., 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17: 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena S., Lenski R. E., 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4: 457–469 [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics: I. Theoretical Introduction. Springer, New York [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Oxford University Press, Oxford [Google Scholar]
- Fogle C. A., Nagle J. L., Desai M. M., 2008. Clonal interference, multiple mutations and adaptation in large asexual populations. Genetics 180: 2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., 2006. Methods for determining spontaneous mutation rates. Methods Enzymol. 409: 195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish P., Lenski R., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102/103: 127–144 [PubMed] [Google Scholar]
- Gresham D., Desai M. M., Tucker C. M., Jenq H. T., Pai D. A., et al. , 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D., 2000. A Primer of Population Genetics. Sinauer Associates, Sunderland, MA [Google Scholar]
- Hegreness M., Kishony R., 2007. Analysis of genetic systems using experimental evolution and whole-genome sequencing. Genome Biol. 8: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M., Shoresh N., Hartl D. L., Kishony R., 1996. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617 [DOI] [PubMed] [Google Scholar]
- Ingolia N. T., Murray A. W., 2007. Positive-feedback loops as a flexible biological module. Curr. Biol. 17: 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. B., Hall D. W., 2004. Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected. Genetics 168: 1817–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao K. C., Sherlock G., 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Murray A. W., 2008. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178: 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Murray A. W., Botstein D., 2009. The cost of gene expression underlies a fitness trade-off in yeast. Proc. Natl. Acad. Sci. USA 106: 5755–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S., Delbrück M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. R., Joyce P., Wichman H. A., 2011. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187: 185–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles R., Gerrish P. J., Moya A., Elena S. F., 1999. Clonal interference and the evolution of RNA viruses. Science 285: 1745–1747 [DOI] [PubMed] [Google Scholar]
- Park S.-C., Krug J., 2007. Clonal interference in large populations. Proc. Natl. Acad. Sci. USA 104: 18135–18140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Simon D., Krug J., 2010. The speed of evolution in large asexual populations. J. Stat. Phys. 138: 381–410 [Google Scholar]
- Peters A. D., Otto S. P., 2003. Liberating genetic variance through sex. BioEssays 25: 533–537 [DOI] [PubMed] [Google Scholar]
- Rosche W. A., Foster P. L., 2000. Determining mutation rates in bacterial populations. Methods 20: 4–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I., Brunet E., Wilke C., 2008. The traveling-wave approach to asexual evolution: Muller’s ratchet and the speed of adaptation. Theor. Popul. Biol. 73: 24–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Ma W. T., Sandri G. H., 1992. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica 85: 173–179 [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G., Lawrence C., 1974. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Wahl L. M., Gerrish P. J., 2001. The probability that beneficial mutations are lost in populations with periodic bottlenecks. Evolution 55: 2606–2610 [DOI] [PubMed] [Google Scholar]
- Wilke C. O., 2004. The speed of adaptation in large asexual populations. Genetics 167: 2045–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]