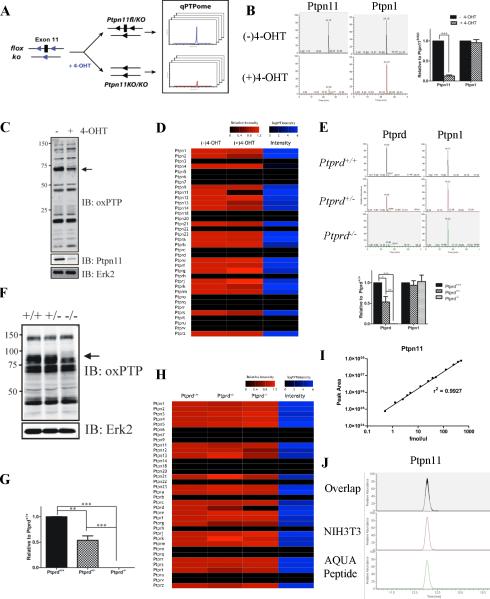
Figure 3. PTP quantification using qPTPome.
(A) Ptpn11fl/KO cells stably expressing CreER were treated with 4-OHT or left untreated, then assessed by qPTPome. (B) Representative SRM profiles for Ptpn11 and Ptpn1. Quantification of each PTP is displayed next to its elution profile (*** p<0.002, paired Student's t-test; 2 biological replicates, each with 2 technical replicates). (C) Immunoblot showing PTP expression levels. Arrow shows position of Ptpn11. (D) Heat map showing relative changes (compared with untreated cells) in PTP expression (measured by qPTPome) following 4-OHT treatment. The intensity value is the average signal for the SRM profiles for each PTP in untreated cells (E) Ptprd+/+, Ptprd+/− and Ptprd−/− brains were profiled by qPTPome (n=3; 3 technical replicates each). Representative SRM profiles are shown for Ptprd and Ptpn1, with quantification of each displayed next to the profiles. Data represent mean ± SEM (* p<0.05; ** p<0.01; *** p<0.001, ANOVA with Bonferroni post-test). (F) Immunoblot with oxPTP Ab showing PTP expression in Ptprd mutant brains. Arrow shows position of Ptprd. (G) Ptprd mRNA levels (assessed by qPCR; n=3). Data represent mean ± SEM (*** p<0.001, ANOVA with Bonferroni post-test). (H) Relative levels of PTP expression (measured by qPTPome) in Ptprd mutant (compared with Ptprd+/+) brains. (I) Standard curve of MS peak area for Ptpn11 AQUA peptide (n=3). (J) SRM peak of the AQUA peptide mixed with native Ptpn11 peptide from NIH3T3 cells. Also, see Figure S3.