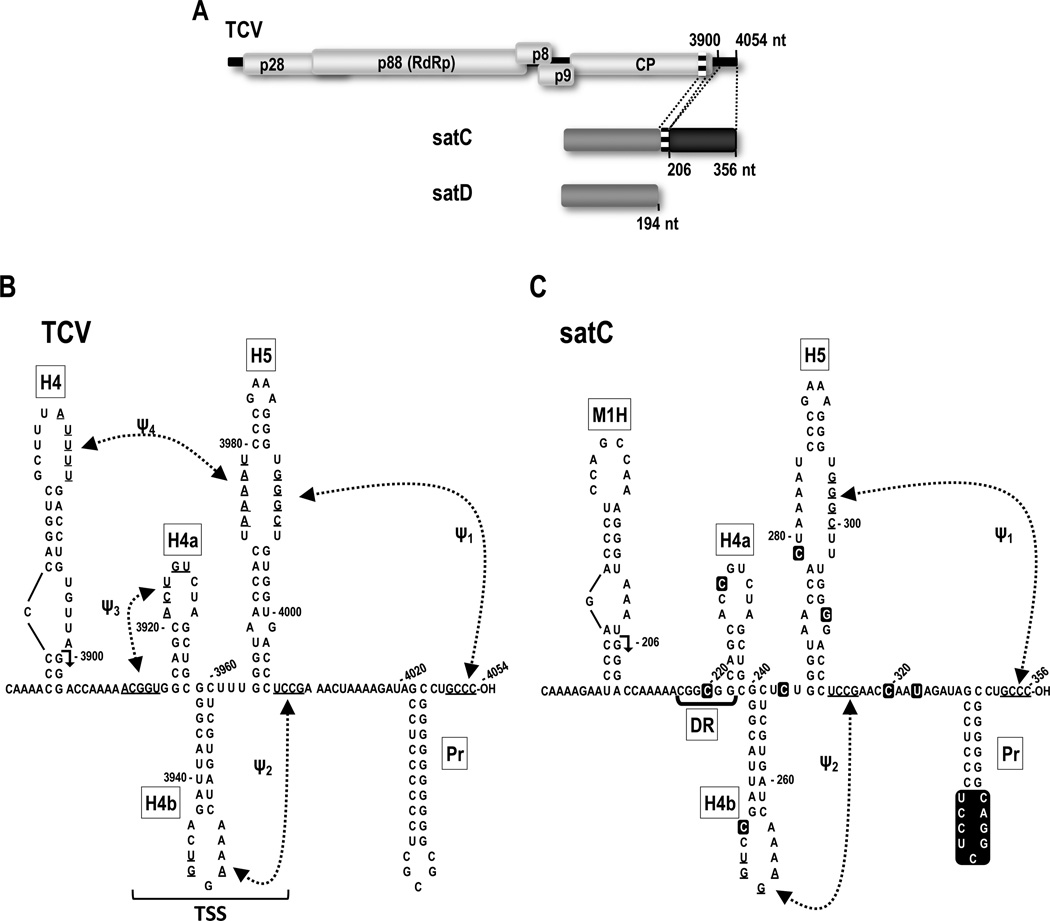
Fig. 1.
Relationship between TCV and satC. (A) Schematic representation of TCV-associated RNAs. The single (+)-strand genome of TCV and five open reading frames are shown. p28 and the read-through protein p88 (the RdRp) are required for replication. p8 and p9 are required for cell-to-cell virus movement. satC is a chimeric RNA composed of a second satRNA (satD) and two regions from the 3' end of TCV. Numbers and dotted lines indicate satC sequences that are shared with TCV. Similar regions are shaded alike. (B) MPGAfold-predicted TCV and (C) satC 3' structures. Hairpins are described in the text. The region the folds into the T-shaped structure (TSS) in TCV is indicated. Sequence differences between satC and TCV are boxed in the satC sequence. Although the satC structure is shown in a similar configuration as TCV for ease of comparison, none of the hairpins are present in the satC structure assumed by transcripts synthesized in vitro, while at least H5 and Ψ1 are present in the satC replication-active structure (Zhang et al., 2004). H4 and M1H are transcriptional enhancers, and H4 is also a critical element for translation (Sun and Simon, 2006; Yuan et al., 2010). Arrowheads in H4 and M1H denote 5' end of shared 3' terminal sequences.