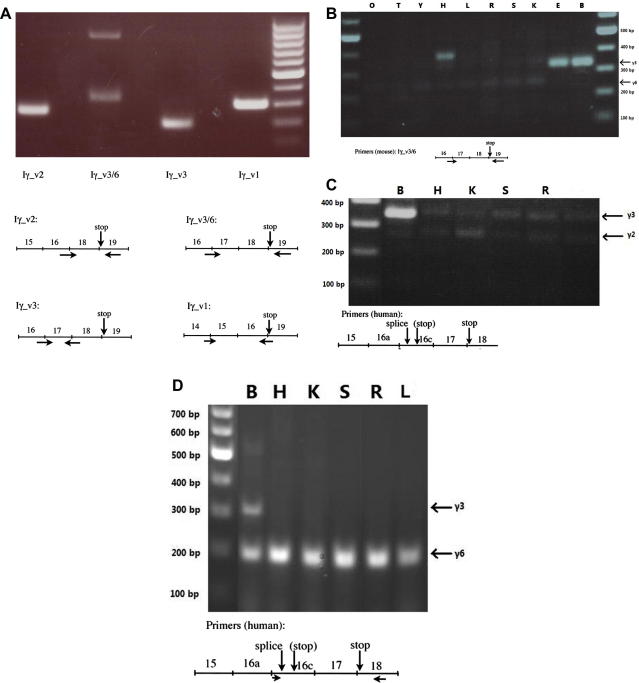
Fig. 2.
PCR amplification of human and rodent cDNA libraries. Primers used for A–D are depicted schematically (not to scale) below each figure. Note the different exon numbering in human versus rodent (see also Fig. 1A). For C and D (human) the alternative splice site within exon 16c that generates Iγ_v3 and Iγ_v6 (see Fig. 1C and text) is indicated by ‘splice’. The stop codon that ends Iγ_v5 (see Ref. [7]) is also indicated in brackets. (A) Rat hippocampal cDNA library [5] using rat-specific primers. (B) Mouse tissues total mRNA library [5] using mouse-specific primers O, ovary; T, testis; Y, thymus; H, heart; L, lung; R, liver; S, spleen; K, kidney; E, embryo; B, brain. The prominent band in heart was sequenced and is not a PIP5KI. (C) Human tissues cDNA library using human-specific primers. B, brain; H, heart; K, kidney; S, spleen; R, liver; L, leukocytes. Note that the lower band in brain and kidney is not Iγ_v6, but Iγ_v2 (see text). (D) Human tissues cDNA library using human-specific primers. For lettering see (C).