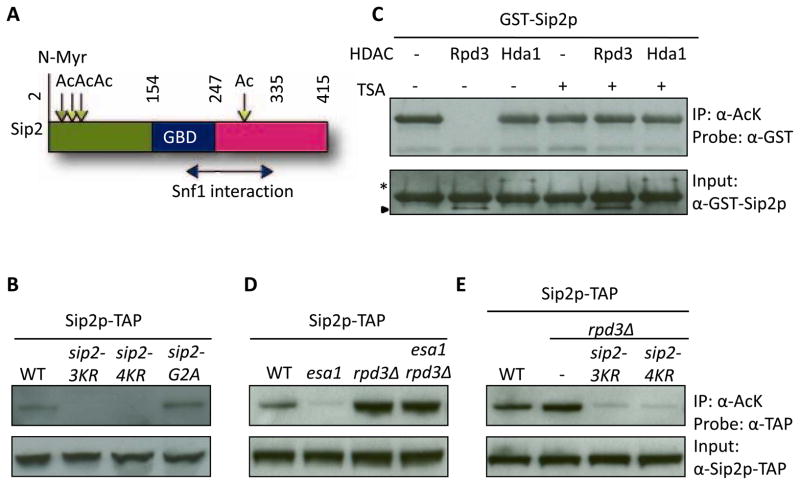
Figure 1. Sip2 Is Acetylated at Four Lysine Sites by Esa1, While Rpd3 Is the Counteracting Deacetylase.
(A) Cartoon of Sip2 structural domains (adapted from Hedbacker, et al. (Hedbacker and Carlson, 2008)). Mass spectrometry identified four acetylated lysine residues of Sip2, K12, K16, K17 and K256. Numbers indicate amino acid residues. Vertical straight line, myristoylation site; Downward arrows, acetylation sites; right-left arrow, regions mapped by deletion analysis as sufficient for Snf1 interaction (Amodeo et al., 2007). Ac, acetylation; N-Myr, N-myristoylation; GBD, glycogen-binding domain.
(B) Chromosomally integrated sip2-3KR and sip2-4KR, but not sip2-G2A mutants, are hypoacetylated in vivo. The sip2-G2A mutant blocks myristoylation. sip2-3KR, sip2-K12/16/17R; sip2-4KR, sip2-K12/16/17/256R.
(C) Rpd3, but not Hda1, removes Sip2 acetylation in vitro. Deacetylation reaction is inhibited by addition of trichostatin A (TSA). Asterisk indicates Hda1-TAP; arrowhead indicates Rpd3-TAP.
(D) Sip2 is hypoacetylated in strains carrying the Ts allele of ESA1, esa1-531, but is hyperacetylated in rpd3Δ; deletion of RPD3 rescues the acetylation defect of esa1-531.
(E) Increased Sip2 acetylation in rpd3Δ is blocked when lysines are mutated to arginines.