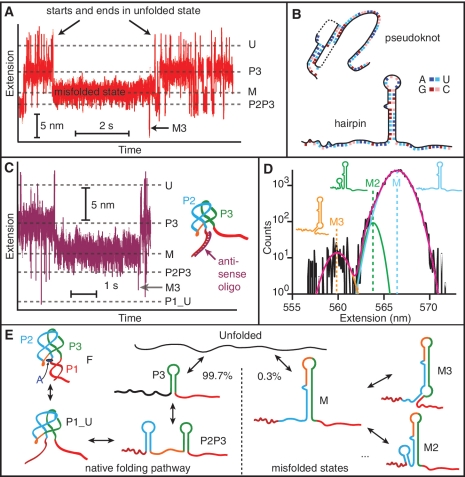
Figure 4.
Misfolded states. (A) Occasionally a state with extension between P3 and P2P3 (labelled ‘M’) forms from the unfolded state. It does not lead to any of the other states on the pathway to the natively folded state F—it begins and ends at U—but it does lead occasionally to other misfolded states with different extensions, such as the state M3. (B) Two potential misfolded structures include a pseudoknot (nucleotides 17–45) and a hairpin (37–68). (C) With a DNA oligomer bound to the 5′ strand of P1, the misfolded state still occurs, indicating that misfolding does not involve the pseudoknot. The P1_U state is still observed even when P1 is prevented from forming. (D) Extension histograms of the off-pathway, misfolded states reveal three distinct misfolded states which are well fit by Gaussian distributions. Insets: structures associated with each state. (E) Folding pathways of the aptamer. The native pathway involves sequential formation of hairpins P3 and P2, then the P2–P3 kissing loop complex (P1_U) and finally the fully-folded aptamer. 0.3% of the time the unfolded state folds into state M, preventing formation of the adenine binding pocket and leading to other off-pathway, misfolded states.