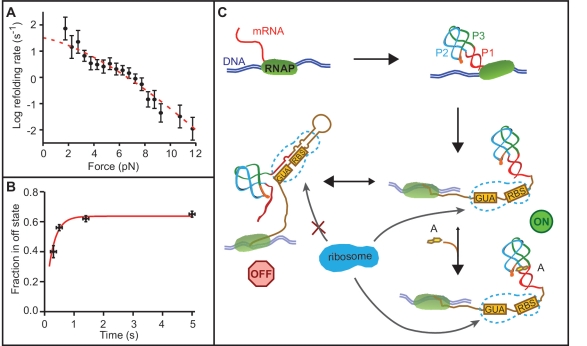
Figure 7.
Folding kinetics and riboswitch mechanism. (A) The aptamer refolding rate as a function of force without adenine is determined from the distribution of refolding forces when ramping the force down from denaturing values. Error bars show SEM. Red line: fit to Equation (2). (B) The fraction of unfolding FECs in the off state as a function of refolding time at low force in the absence of adenine shows an exponential rise as the riboswitch structure equilibrates into the more stable ‘off’ state. (C) Schematic of the riboswitch mechanism. The aptamer folds rapidly before the expression platform is transcribed, regardless of adenine binding. If adenine binds to the aptamer, it stabilizes the ‘on’ state of the riboswitch (aptamer folded, ribosome binding site exposed). Without adenine binding, the ‘on’ state is unstable and equilibrates into the ‘off’ state (P1 unfolded, ribosome binding site sequestered). RBS: ribosome binding site.