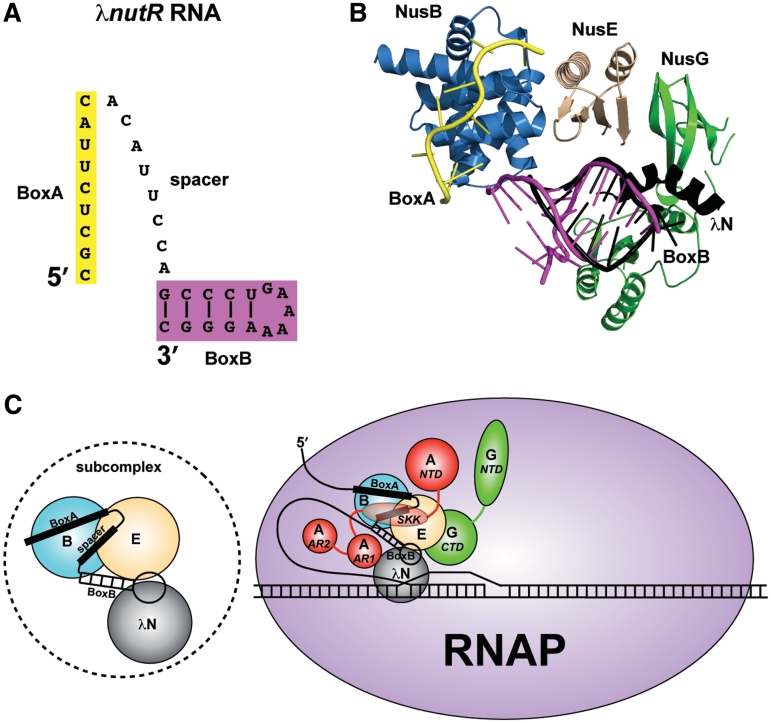
Figure 6.
Models for the λN-mediated antitermination subcomplex and complete complex. (A) Representation of the λnutR RNA showing the possible spatial relationship between BoxA (yellow) and BoxB (magenta) components. (B) Superimposed components of the antitermination complex with known structures, including AaNusB–NusE–BoxA and AaNusB–NusE–dsRNA (this work), the N-terminal α-helix of λN protein bound to BoxB (PDB ID: 1QFQ, black), the C-terminal domain of EcNusG in its complex with EcNusE (PDB ID: 2KVQ, green), and the N-terminal domain of EcNusG (PDB ID: 2K06, green). The N- and C-terminal domains of EcNusG were positioned relative to one another by superimposing them with the N- and C-terminal domains, respectively, of AaNusG (PDB ID: 1NPP). (C) Left: the NusB–NusE–BoxA–spacer–BoxB–λN subcomplex; Right: updated model for the complete λN-mediated antitermination complex, illustrating all known interactions. Nus factors B (blue) and E (wheat) cooperatively bind BoxA and BoxB (this work). λN protein (gray) binds BoxB (8–11) and also possibly NusE (this work). λN protein binds RNAP near the transcription bubble (45). NusA (red) stabilizes these interactions by interacting with the λnut spacer RNA between BoxA and BoxB (12), and with λN protein (51) through NusA SKK (S1-KH1-KH2) and AR1 domains, respectively, while anchoring the complex to RNAP through the NusA NTD and AR2 domains (48–50). NusG (green) further stabilizes the complex through its CTD interaction with NusE (5), also serving as an anchor to RNAP through its NTD (53).