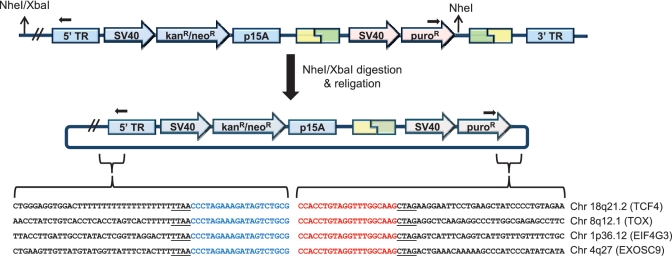
Figure 6.
Mapping the chromosomal locations of donor plasmid integrations into the transposon target site. Genomic DNA of the polyclonal G418R/puroR HEK293 cell populations was digested with NheI and XbaI, circularized by ligation and transformed into E. coli. Purified plasmid was sequenced by reverse and forward primers that bind to the transposon and donor plasmid, respectively, to ensure recovery of correctly targeted transposons. The recovered sequences were intragenic, which is a favored location for piggyBac transposition (8). Gene names and chromosomal locations are indicated to the right of the corresponding sequencing results. The positions of sequencing primers are indicated with black arrows. The underlined TTAA and CTAG correspond to the site of piggyBac transposition and the NheI/XbaI compatible 5′ overhang, respectively.