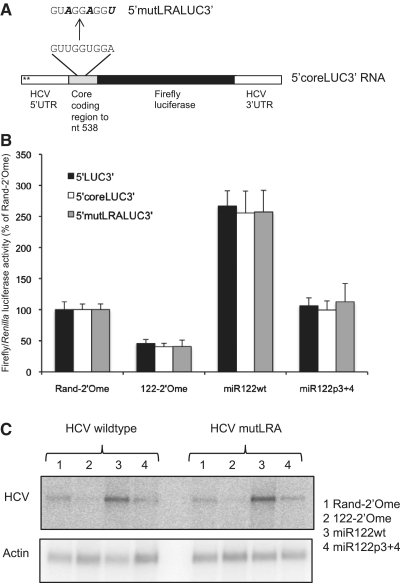
Figure 6.
miR-122 does not activate translation by disruption of a proposed long-range annealing (LRA) motif. (A) Part of the HCV core protein coding region, encompassing the sequence predicted to form an LRA with the miR-122 binding site, was fused in frame to the HCV IRES in p5′LUC3′ to generate a plasmid encoding 5′coreLUC3′ RNA. The 5′mutLRALUC3′ RNA has three point mutations that disrupt the LRA while maintaining the amino acid sequence. (B) 5′coreLUC3′ and 5′mutLRALUC3′ RNA were introduced into Huh7 cells with sequestration or overexpression of miR-122. Firefly luciferase activity is shown relative to Renilla luciferase activity of the transfection control as a percentage of the Rand-2′Ome values for each RNA. Data are an average of three independent triplicate experiments and error bars represent standard deviation. The responses of 5′coreLUC3′ and 5′mutLRALUC3′ to 122-2′Ome or miR122wt did not differ significantly to that of 5′LUC3′ RNA. (C) The mutation to the LRA shown in (A) was introduced into a plasmid encoding replication-competent H77ΔE1/p7 HCV RNA. Wild-type and mutant RNA were introduced into Huh7 cells and HCV RNA levels after 5 days were determined by northern blotting following sequestration or overexpression of miR-122. γ-actin RNA is shown as a loading control. The LRA mutation did not affect HCV RNA abundance or the response to miR-122.