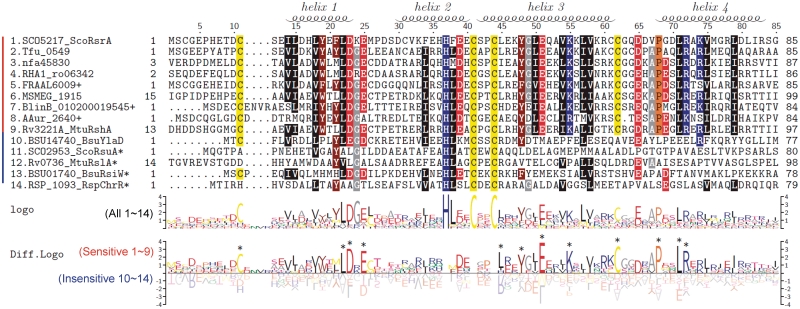
Figure 3.
Multiple sequence alignment and sequence logo of ScoRsrA homologs. Mulitple sequence alignment was carried out by using MEGA4, followed by manual curation. Sequence logos were produced by using TEXshade. The secondary structure prediction is based on the structural information of RspChrR (PDB;2Z2S). For alignment, we trimmed both N-terminal and C-terminal residues that extend beyond the ScoRsrA sequence. The nucleotide sequencing identified some sequence differences from the public database, and we used modified sequences for BlinB_010200019545, AAur_2640 and FRAAL6009 (+; Supplementary Table S3). Asterisk indicates entries that contain C-terminal domains in addition to ASD.