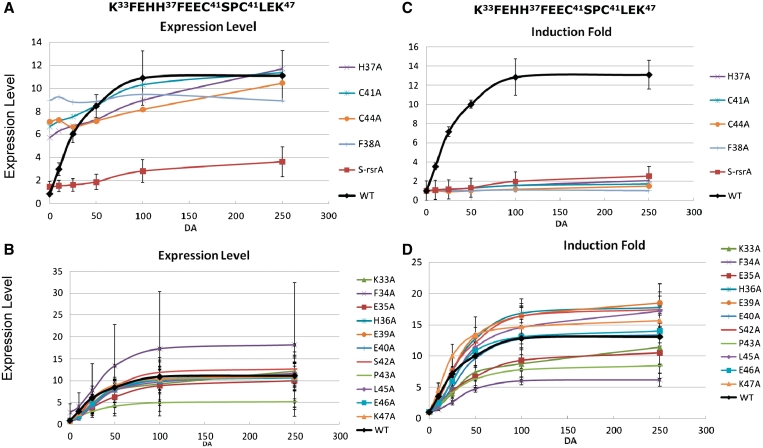
Figure 5.
Redox sensitivies of Ala-scanned ScoRsrA mutants. Each of the 15 residues from K33 to K47 in ScoRsrA was changed to alanine in recombinant S. coelicolor strains that harbor mutated rsrA in sigR-rsrA operon inserted at the att site of the ΔsigR-rsrA strain. Cells were grown to OD600 of ∼0.3 in YEME and treated with varying concentrations of diamide (0, 10, 25, 50, 100, 250 μM) for 10 min. Following S1 mapping, the normalized sigRp2 signal values were plotted either as expression levels (A and B) or induction fold (C and D) against diamide concentrations. For expression values, the uninduced basal level of the wild-type was set as 1.0. For induction folds, the uninduced levels of all strains were set as 1.0. Mutants that constitutively induced SigR-specific expression were presented separately (panels A and C) from those that affected primarily sensitivity to diamide (panels B and D). Quantified values for induction folds were presented in Table 1.