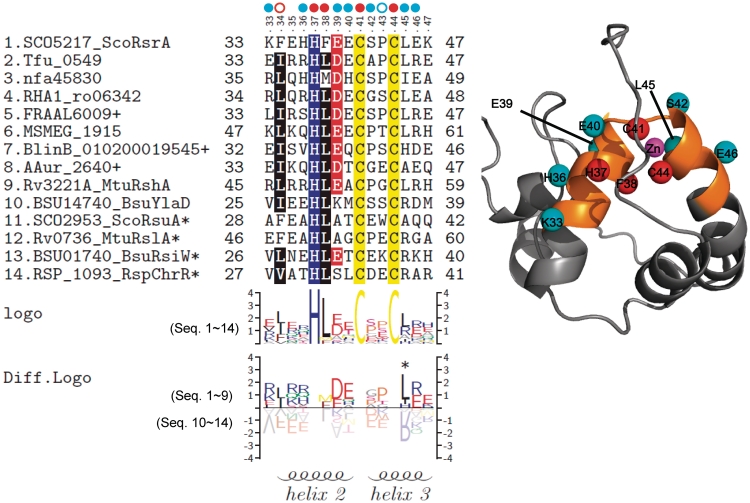
Figure 6.
The conserved sequence pattern and the position of the residues near the HCC motif in 3D structural model of ScoRsrA. Multiple sequence alignment and sequence logos for the 15 amino acid region examined for mutational analyses were presented as in Figure 3. A 3D structural model was produced through homology modeling, using RspChrR structure (PDB; 2Z2S) as a template. Residues that affected redox-sensitivity (IDIF50) by more than 2-fold were indicated with cyan circles, whereas those that affected structural integrity of ScoRsrA were indicated with red ones. Mutations that partially affected ScoRsrA structure by decreasing interaction with SigR (F34, red open circle) or increasing interaction with SigR (P43, blue open circle) were also indicated. In the structure model, Cβ atoms of effective residues were highlighted as spheres with indicated colors. The mutated 15 amino acid region in 3D structure was highlighted in orange color.