Figure 4.
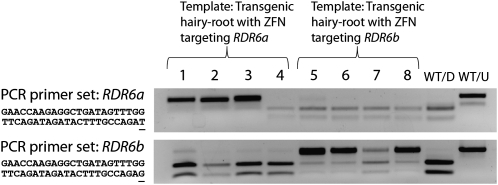
The mutagenic specificity of the RDR6a and RDR6b ZFN transgenes was assessed by performing PCR enrichment assays with gene-specific primers for each homeolog. The ZFN target sites of this gene pair differ by only two nucleotides, so this experiment was important to measure whether the gene-specific ZFNs could discriminate between the homeologous targets (Table I provides details on the target site differences of the two ZFN transgenes). The PCR enrichment assays are analogous to those shown in Figure 2. A sample of estradiol-induced hairy roots was targeted for mutagenesis using the ZFN transgene targeting RDR6a (roots 1–4) and the ZFN transgene targeting RDR6b (roots 5–8). Primer sets differing at a single nucleotide were designed to allow for homeolog-specific PCR amplification in these samples (the polymorphic nucleotide is underlined in the reverse primer sequences). The top section shows the PCR enrichment results when testing for mutagenesis of gene RDR6a and the bottom section shows the PCR enrichment results when testing for mutagenesis of gene RDR6b. In either case, an undigested top band indicates a mutation within the hairy-root sample. A digested nontransgenic root sample (WT/D) and an undigested nontransgenic root sample (WT/U) serve as controls. Seven out of the eight targeted hairy roots show the presence of the putative mutated top band; root 4 does not show this band, indicating that this sample either failed to transform or the ZFN failed to mutagenize any cells in this root. Faint top band shadows are observed in some samples for the nontargeted gene. Therefore, we cannot rule out the possibility that some mutations may have occurred at the nontargeted homeologous gene, albeit at a much lower frequency than the targeted gene.