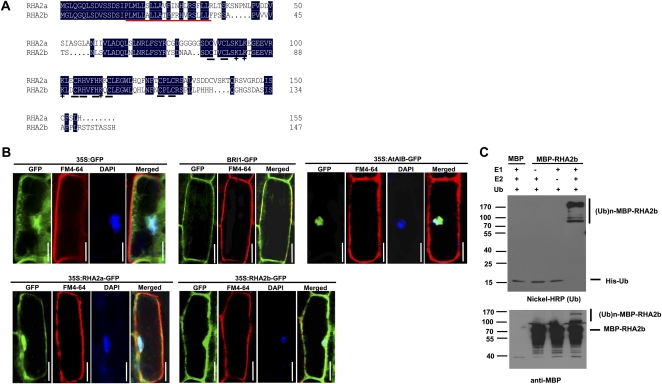
Figure 1.
RHA2b exhibits E3 ligase activity. A, Alignment of deduced amino acid sequences of RHA2b and RHA2a. Conserved residues are highlighted in black, and Cys and His residues in the RING finger domain are underlined in black. The putative transmembrane domain is underlined in red, and basic amino acids in the RING finger domain are marked by cross stars. B, Subcellular localization of RHA2b and RHA2a. For 35S:GFP, 35S:AtAIB-GFP, 35S:RHA2a-GFP, or 35S:RHA2b-GFP, roots of 1-week-old seedlings were immersed in 2 μg mL−1 DAPI solution for 10 to 15 min and then stained with 5 μg mL−1 cold FM4-64 solution for 1 min. Before staining, the whole seedlings of 35S:RHA2a-GFP or 35S:RHA2b-GFP were pretreated with 50 μm MG132 for 6 h under dim-light conditions; for BRI1-GFP, roots of 1-week-old seedlings were stained with 5 μg mL−1 cold FM4-64 solution for 1 min. All seedlings were observed with fluorescence microscopy. Bars = 30 μm. C, MBP-RHA2b fusion protein was assayed for E3 activity in the presence of E1 (from wheat [Triticum aestivum]), E2 (UBCh5b), and 6× His tag ubiquitin (Ub). The numbers at left denote the molecular masses of marker proteins in kD. MBP itself was used as a negative control. Samples were resolved by 8% SDS-PAGE. Nickel-horseradish peroxidase (HRP) was used to detect His tag ubiquitin (top panel), and the anti-MBP antibody was used for maltose fusion proteins (bottom panel).