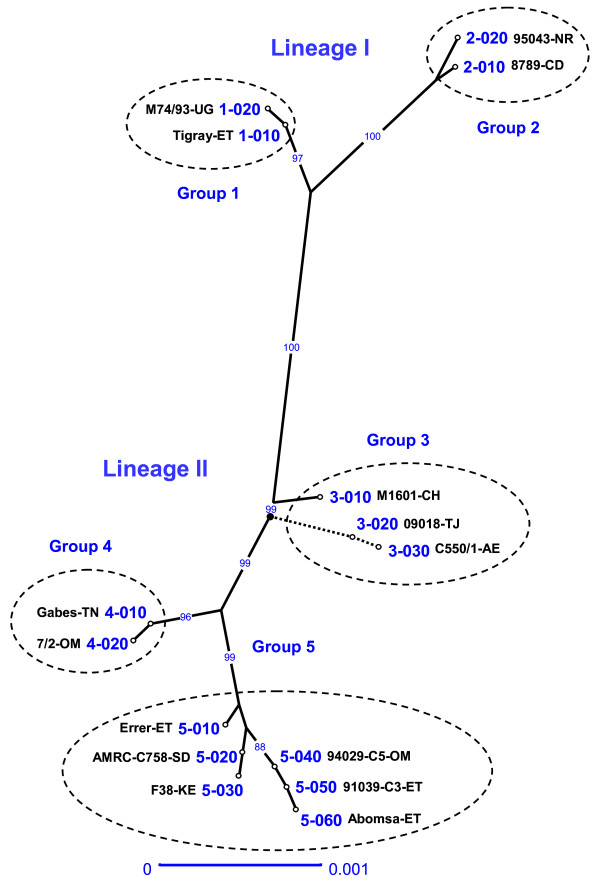
Figure 1.
Tree derived from distance analysis of the eight concatenated MLSA loci. The tree was constructed using the unweighted neighbour-joining algorithm (Darwin 5.0). A single strain representing each of the 15 MLSA ST is displayed (see Table 1 for strain details). The two sequences presenting a large 960 nt deletion (09018 and C550/1) were grafted at their respective positions after tree construction (discontinuous lines) in order to avoid their influence during tree inference. The root of the tree is represented as a bold dot. Bootstrap percentage values were calculated from 1000 resamplings and values over 80% are displayed. The scale bar shows the equivalent distance to 1 substitution per 1000 nucleotide positions. The ST were numbered according to the group in which they clustered, followed by a three digit code that should allow intercalating additional ST as new sequences are obtained. Note that the two letter code following the strain name represents the corresponding country of isolation: AE, United Arab Emirates; CD, Chad; CH, China; ET, Ethiopia; NR, Niger; OM, Oman; SD, Sudan; TJ, Tajikistan; TN, Tunisia; UG, Uganda.