Abstract
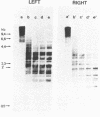
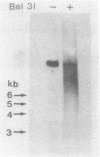
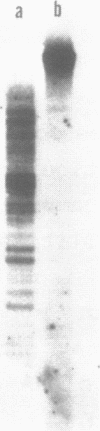
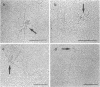
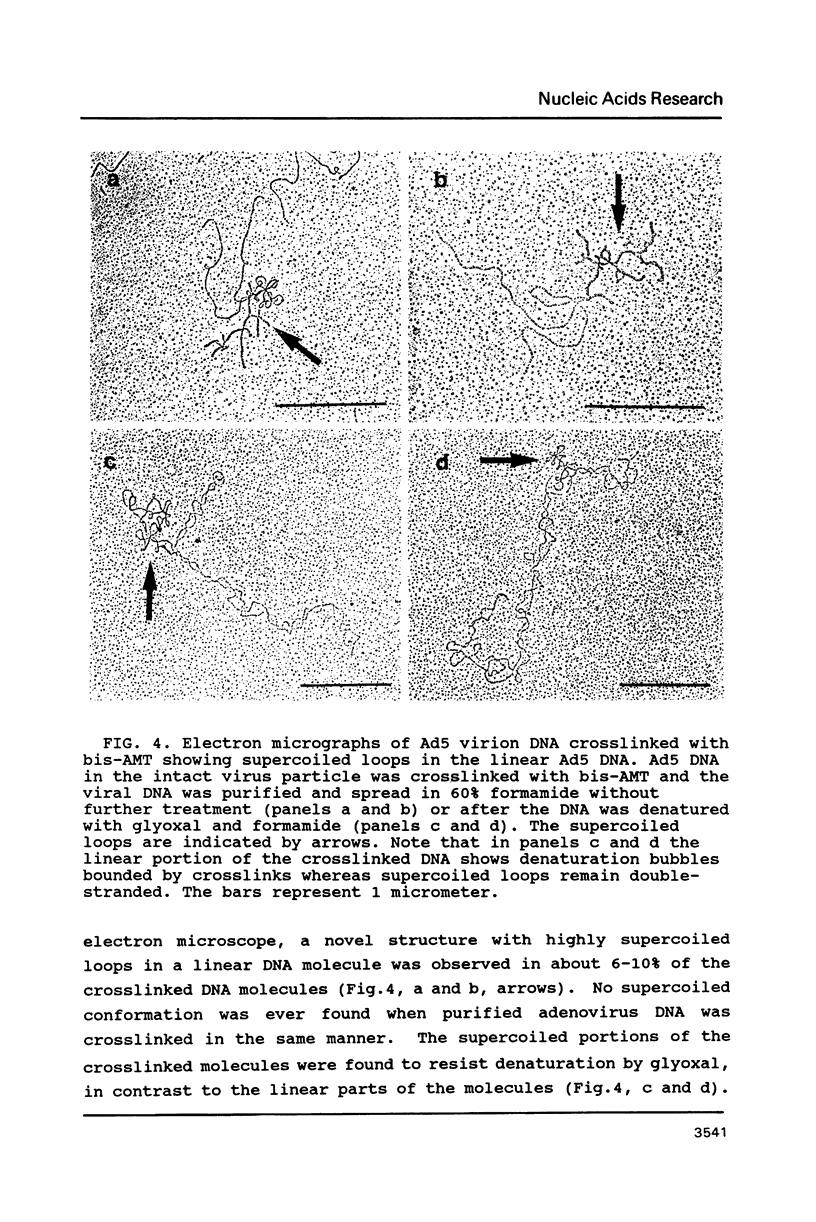
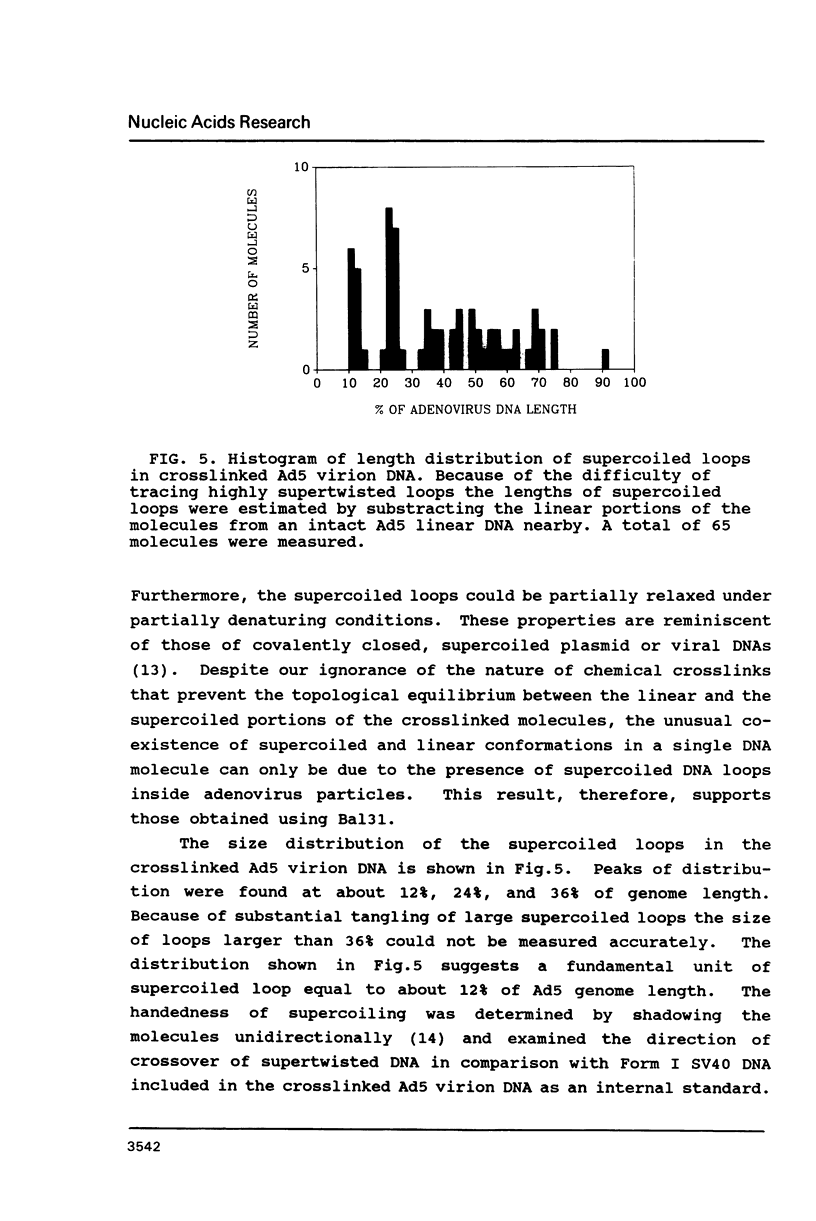
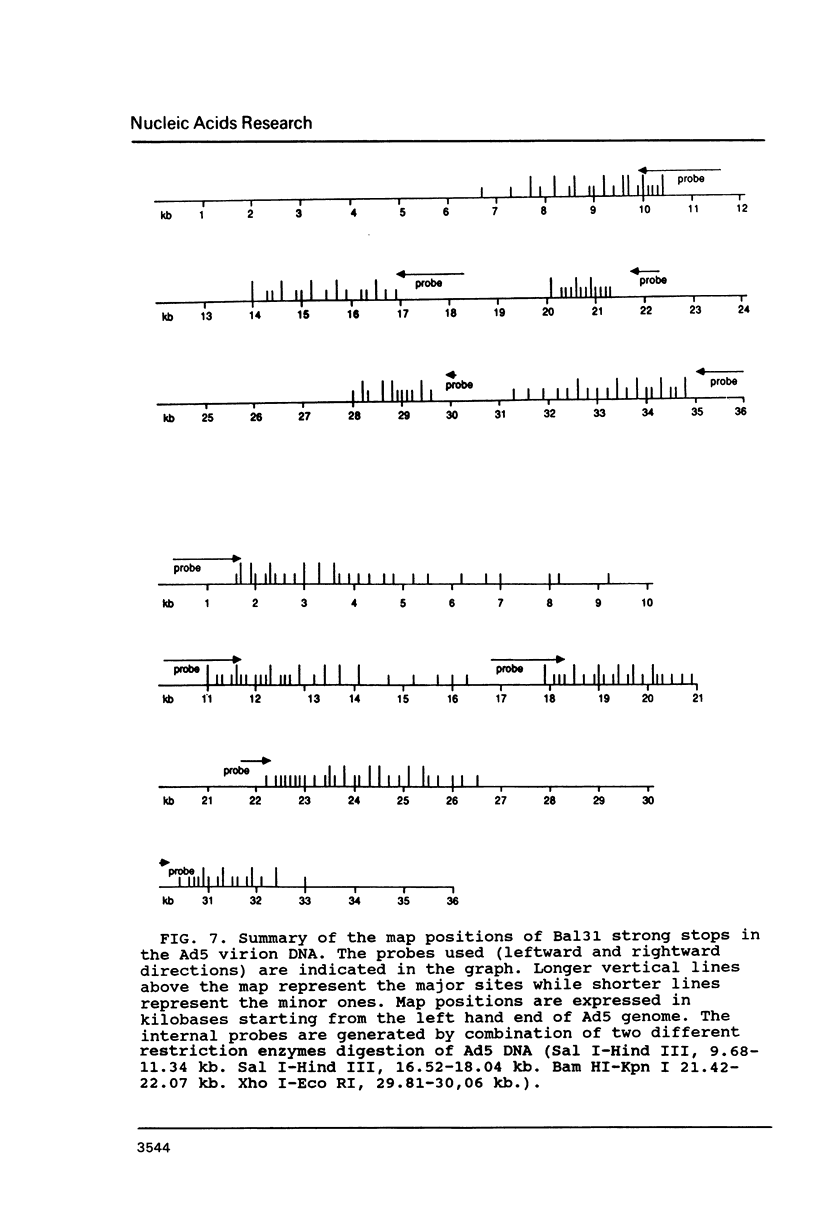
Electron microscopic analysis of bis-psoralen crosslinked adenovirus type 5 virion DNA revealed supercoiled domains in an otherwise linear DNA. The existence of supercoiled arrangement in all the virion DNA was demonstrated by the sensitivity of Ad5 DNA in pentonless virus particles to the supercoiling-dependent endonucleolytic activity of Bal31 and S1 nucleases. These nucleases were found to cleave Ad5 virion DNA at specific sites. The observation of stable cleavage sites in the limit digestion of virion DNA by Bal31 suggests that cleavage sites represent boundaries of core proteins which impede the exonuclease activity of Bal31. These data suggest that specific arrangement of core proteins on Ad5 virion DNA. Based on this analysis we determined positions of core proteins in viral genome using indirect end labeling technique. The size of supercoiled domains of virion DNA was estimated by electron microscopy and also by boundaries of mutually exclusive Bal31 cleavage sites at limit digestion condition. Our data suggest each supercoiled domain is equal to about 12% of Ad5 genome length and about 8 loops can be accommodated in Ad5 virion. However sequences at two extreme ends of the viral genome were found to be outside of supercoiled domains. An interesting correlation between supercoiled domains and gene domains of Ad5 genome was noticed.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broker T. R., Soll L., Chow L. T. Underwound loops in self-renatured DNA can be diagnostic of inverted duplications and translocated sequences. J Mol Biol. 1977 Jul 15;113(4):579–589. doi: 10.1016/0022-2836(77)90223-6. [DOI] [PubMed] [Google Scholar]
- Brown D. T., Westphal M., Burlingham B. T., Winterhoff U., Doerfler W. Structure and composition of the adenovirus type 2 core. J Virol. 1975 Aug;16(2):366–387. doi: 10.1128/jvi.16.2.366-387.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino G. D., Gamper H. B., Isaacs S. T., Hearst J. E. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- Corden J., Engelking H. M., Pearson G. D. Chromatin-like organization of the adenovirus chromosome. Proc Natl Acad Sci U S A. 1976 Feb;73(2):401–404. doi: 10.1073/pnas.73.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto S., Hsu M. T. Determination of twist and handedness of a 39-base pair segment of DNA in solution. Nature. 1983 Sep 1;305(5929):70–72. doi: 10.1038/305070a0. [DOI] [PubMed] [Google Scholar]
- Lau P. P., Gray H. B., Jr Extracellular nucleases of Alteromonas espejiana BAL 31.IV. The single strand-specific deoxyriboendonuclease activity as a probe for regions of altered secondary structure in negatively and positively supercoiled closed circular DNA. Nucleic Acids Res. 1979 Jan;6(1):331–357. doi: 10.1093/nar/6.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M. A., Weber J. Structure of adenovirus chromatin. Biochim Biophys Acta. 1982 Jan 26;696(1):76–86. doi: 10.1016/0167-4781(82)90012-4. [DOI] [PubMed] [Google Scholar]
- Nermut M. V. Structural elements in adenovirus cores. Evidence for a "core shell" and linear structures in "relaxed" cores. Arch Virol. 1979;62(2):101–116. doi: 10.1007/BF01318063. [DOI] [PubMed] [Google Scholar]
- Nermut M. V. The architecture of adenoviruses: recent views and problems: Brief review. Arch Virol. 1980;64(3):175–196. doi: 10.1007/BF01322699. [DOI] [PubMed] [Google Scholar]
- Newcomb W. W., Boring J. W., Brown J. C. Ion etching of human adenovirus 2: structure of the core. J Virol. 1984 Jul;51(1):52–56. doi: 10.1128/jvi.51.1.52-56.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W. A., Walter C. F., Cryer B. L. Barriers to nuclease Bal31 digestion across specific sites in simian virus 40 chromatin. Mol Cell Biol. 1984 Apr;4(4):604–610. doi: 10.1128/mcb.4.4.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Carlson J. O., Pettijohn D. E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980 Oct;21(3):773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Torsional tension in intracellular bacteriophage T4 DNA. Evidence that a linear DNA duplex can be supercoiled in vivo. J Mol Biol. 1982 Dec 15;162(3):659–677. doi: 10.1016/0022-2836(82)90394-1. [DOI] [PubMed] [Google Scholar]
- Vayda M. E., Rogers A. E., Flint S. J. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983 Jan 25;11(2):441–460. doi: 10.1093/nar/11.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965 May;53(5):1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virrankoski-Castrodeza V., Parish J. H. Evidence for supercoiling in the DNA of bacteriophage heads. Arch Microbiol. 1980 Jul;126(3):277–283. doi: 10.1007/BF00409932. [DOI] [PubMed] [Google Scholar]
- Wilhelm J., Brison O., Kedinger C., Chambon P. Characterization of adenovirus type 2 transcriptional complexes isolated from infected HeLa cell nuclei. J Virol. 1976 Jul;19(1):61–81. doi: 10.1128/jvi.19.1.61-81.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth D. J., Hsu M. T. Visualization of nascent RNA transcripts and simultaneous transcription and replication in viral nucleoprotein complexes from adenovirus 2-infected HeLa cells. J Mol Biol. 1981 Apr 5;147(2):247–268. doi: 10.1016/0022-2836(81)90440-x. [DOI] [PubMed] [Google Scholar]
- Wu C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature. 1984 Sep 6;311(5981):81–84. doi: 10.1038/311081a0. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]