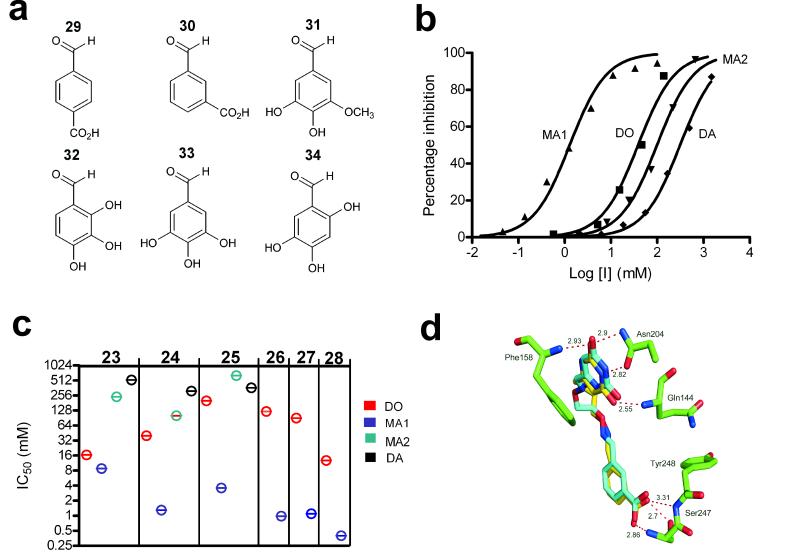
Figure 3.
Structures and inhibition profiles of library aldehyde fragments containing DO, MA1, MA2 and DA linkers. (a) Structure of library aldehydes used in the synthesis of DO, MA1, MA2 and DA libraries. (b) Concentration dependence of hUNG2 inhibition by the amine and oxime linker compounds generated from library aldehyde 30. The IC50 values were DO (14) = 40 μM; MA1 (6) = 1.3 μM; MA2 (22) = 100 μM; DA (27) = 315 μM (c) IC50 values for library aldehyde fragments 29 – 34 in the context of DO, MA1, MA2 and DA linkages. For aldehyde fragments 32 – 34, only DO and MA1 linkers were tested. Experiments were repeated in triplicate and errors are standard deviations of the data from the fitted curve. (d) Conformations and interactions of the bound DO (14) and MA1 (6) inhibitors derived from library aldehyde 30 (see Supplemental Figure 1 for electron density map of the complex with 6). The structures of MA2 (22) and DA (27) revealed that, for these compounds, fragment 30 did not interact with its binding site (Supplemental Figure 2 online).