Abstract
Genes of the major histocompatibility complex (MHC) have a crucial role in the immune response of vertebrates, alter the individual odour and are involved in shaping mating preferences. Pathogen-mediated selection, sexual selection and maternal–fetal interactions have been proposed as the main drivers of frequently observed high levels of polymorphism in functionally important parts of the MHC. Bats constitute the second largest mammalian order and have recently emerged as important vectors of infectious diseases. In addition, Chiroptera are interesting study subjects in evolutionary ecology in the context of olfactory communication, mate choice and associated fitness benefits. Thus, it is surprising that they belong to the least studied mammalian taxa in terms of their MHC diversity. In this study, we investigated the variability in the functionally important MHC class II gene DRB, evidence for selection and population structure in the group-living lesser bulldog bat, Noctilio albiventris, in Panama. We found a single expressed, polymorphic Noal-DRB gene. The substitution pattern of the nucleotide sequences of the 18 detected alleles provided evidence for positive selection acting above the evolutionary history of the species in shaping MHC diversity. Roosting colonies were not genetically differentiated but females showed lower levels of heterozygosity than males, which might be a sign that the sexes differ in the selection pressures acting on the MHC. This study provides the prerequisites for further investigations of the role of the individual MHC constitution in parasite resistance, olfactory communication and mate choice in N. albiventris and other bats.
Keywords: major histocompatibility complex polymorphism, selection, population structure, neotropical bat, Noctilio albiventris, Panama
Introduction
Genes of the major histocompatibility complex (MHC) play an essential role in the adaptive immune response of vertebrates and are crucial for the understanding of the influence of natural selection on genetic diversity in wild populations (Bernatchez and Landry, 2003). MHC-encoded cell surface glycoproteins present antigens of intracellular (MHC class I genes) and extracellular (MHC class II genes) origin to T cells that trigger the appropriate immune response (Klein, 1986; Janeway and Travers, 2002; Kumanovics et al., 2003). Class II-derived molecules are heterodimers of two associated polypeptide chains, an α- and a β-chain. The β-chain consists of an antigen-binding β1-domain, an immunoglobulin-like β2-domain and a transmembrane domain. Certain amino-acid positions of the β1-region, the so-called antigen-binding sites (ABS, encoded by exon 2), show high levels of genetic variation (Brown et al., 1993) with a higher rate of non-synonymous (dN, amino-acid altering) over synonymous (dS, silent) nucleotide substitutions (Hughes and Nei, 1988, 1989). According to neutrality theory (Kimura, 1977), the rate of synonymous nucleotide substitution is predicted to be larger than the rate of non-synonymous substitutions because a change of the amino-acid composition is more likely to be deleterious. The pattern of an elevated rate of non-synonymous substitutions at ABS is considered as clear evidence for ‘positive Darwinian selection' (Hughes and Nei 1988, 1989) shaping genetic variation (reviewed in Hughes, 2007). Others used the term ‘historical positive selection' (Hedrick, 1999) or ‘positive selection over evolutionary time scale' (Bernatchez and Landry, 2003) to describe this observation. This polymorphism enables the immune system to recognise an extensive range of pathogens and is therefore crucial for the immunological fitness of individuals and, thus, animal populations (Edwards and Potts, 1996; Bernatchez and Landry, 2003). The high levels of polymorphism at MHC class II loci found in most vertebrate species are thought to be maintained by different forms of balancing selection, including heterozygosity advantage (Doherty and Zinkernagel, 1975) and rare allele advantage (also called negative frequency-dependent selection; Takahata and Nei, 1990; Slade and McCallum, 1992) as well as selection that varies in space and time (Hill, 1991; Hedrick, 2002). Distinguishing between the different forms of balancing selection in natural populations is difficult, as they are not mutually exclusive and may operate together with other selective and neutral forces (Sommer, 2005; Piertney and Oliver, 2006; Spurgin and Richardson, 2010).
Genes of the MHC are known to alter the individual body odour (for example, Penn and Potts, 1998; Milinski, 2006; Kwak et al., 2009) and may therefore be involved in mate choice decisions (for example, Wedekind et al., 1995; Chaix et al., 2008; Ilmonen et al., 2009), thus promoting genetic structure within and among social groups (Chesser, 1990; Kundu and Faulkes, 2004; Matocq and Lacy, 2004; Cutrera and Lacey, 2006). Sexual selection processes may not only be supported in direct mate choice situations but also through postcopulatory mechanisms including maternal–fetal interactions (for example, Wedekind et al., 1996; Ober and Van der Ven, 1997). Several studies have already highlighted that reproduction among MHC-dissimilar mates is favoured (for example, primates: Schwensow et al., 2008; and humans: Chaix et al., 2008; Ilmonen et al., 2009); mostly because a genetically diverse offspring genotype is advantageous in the defence against pathogens (Parham and Otha, 1996; Penn and Potts, 1999). Yet, the debate is still controversial. In some populations, MHC-based mate choice was not affirmed (humans: Hedrick and Black, 1997; Chaix et al., 2008; and ruminants: Patterson and Pemberton, 1997). Some studies have suggested that an intermediate rather than the highest level of MHC diversity is optimal (Wegner et al., 2003; Woelfig et al., 2009).
Bats are not only the second largest mammalian order but also the most gregarious of all mammals. Some bat colonies harbour several million individuals. Thus, social communication among bats can be complex, involving not only visual and acoustical but also olfactory signals (McCracken and Wilkinson, 2000; Altringham and Fenton, 2003). Many bat species produce distinct odours from a variety of glands (for example, Quay, 1970; Dapson et al., 1977; Voigt and von Helversen, 1999; Scully et al., 2000; Caspers et al., 2009). Pilot studies have shown that bats use odours for kin and individual recognition (Gustin and McCracken, 1987; Bouchard, 2001; Safi and Kerth, 2003) and during male–female interactions (Voigt and von Helversen, 1999; Bouchard, 2001). Recently, it has been discovered that bats are reservoir hosts and potentially important vectors of many infectious diseases (Calisher et al., 2006; Wong et al., 2007; Field, 2009; Wibbelt et al., 2010). Given the combination of these facts, it becomes apparent that the study of bat immunogenetics is of fundamental relevance not only for understanding the mechanisms underlying the zoonotic potential of bats but also to shed light on the link between MHC, pathogen resistance and olfaction-based mate choice in mammals.
In the present study, we examined the MHC class II DRB region of a neotropical bat species, the lesser bulldog bat, Noctilio albiventris, in Panama. N. albiventris lives in social groups of so far unknown relatedness. Besides acoustical signals, group members also communicate among each other via volatile compounds from the so-called oily spots in the subaxillary region. In addition, males possess inguinal pockets next to the scrotum that provide a distinct odour, particularly during the reproductive period (Studier and Lavoie, 1984; Brook and Decker, 1996). In summary, N. albiventris harbours morphologically distinct scent-producing organs, suggesting that volatiles are of key importance not only for individual communication but also for mate choice.
Our specific aims were: (1) to design reliable primers to characterise MHC class II DRB exon 2 diversity in N. albiventris, using a genome walking approach that enables the amplification of specific DNA fragments in situations where the sequence of only one primer is known (modified from Ko et al., 2003), (2) to examine evidence for selection acting on MHC in N. albiventris, that is, a higher rate of non-synonymous over synonymous substitutions in ABS would indicate positive selection acting above the evolutionary history of the species and (3) to investigate whether subpopulations, roosting colonies and the sexes differ in their genetic population structure. Differences in the MHC constitution of subpopulations and roosting colonies could indicate local adaptation as a result of recent selection events, whereas differences between sexes could indicate MHC-related sexual selection mechanisms.
Materials and methods
Study site and sampling
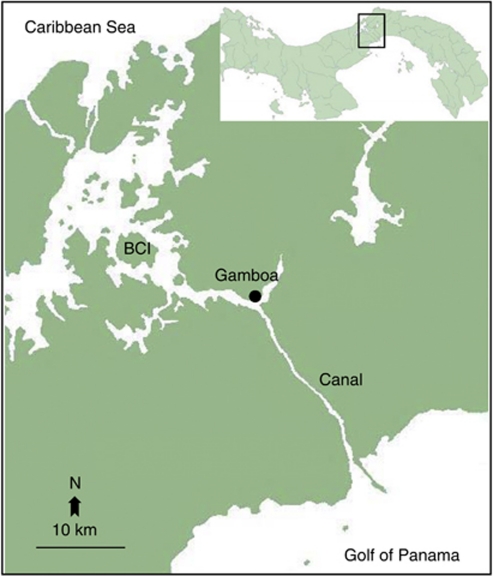
Our main study site was the village Gamboa (09.07° N and 079.41° W) in Panama. Bats were caught with mist nets (see Dechmann et al., 2009) as they emerged at dusk from their roost during 2006–2008. Individuals caught from the same daytime roost were assumed to be from the same colony. A second study site was Barro Colorado Island (BCI; 09.10° N and 079.51° W) situated 15 km away from Gamboa, where animals foraging above water were caught in mist nets set up along the boat docks of a marina (Figure 1). Bats from the two sites were considered separate subpopulations as their foraging areas did not overlap (Dechmann et al., 2009). Bats were sexed, aged and body measurements taken. From all 215 bats, we collected a 4-mm skin sample from the wing membrane using a sterile biopsy punch (Worthington Wilmer and Barrat, 1996). Skin samples were stored in 96% ethanol until DNA isolation.
Figure 1.
Map of the study sites Gamboa and BCI in Panama (modified from the Smithonian Tropical Research Institute mapserver).
For optimal primer design and to prove expression of MHC alleles, a liver sample was collected from an euthanised male N. albiventris. The liver sample was preserved in RNA later (Sigma-Aldrich, Steinheim, Germany) and stored at −20 °C until subsequent analysis.
Genomic DNA and RNA isolation and complementary DNA library construction
Genomic DNA (gDNA) was extracted using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. RNA was isolated from the liver using the RNeasy Kit (Qiagen) that includes a DNase treatment according to the manufacturer's instructions. The quantity was determined by measuring the absorbance at 260 nm and purity (that is presence of contaminants) was assessed by the ratio at 260/280 nm, which should be ∼1.8 for DNA and ∼2.0 for RNA (Nanotrop, Peqlab, Erlangen, Germany). Up to 5 μg of total RNA were used to construct a first strand complementary DNA (cDNA) library. For reverse transcription, we included 5 μg Oligo(dT)12–18 Primer (Invitrogen, Karlsruhe, Germany) and 200 U of Revert Aid H-Minus M-MulV reverse transcriptase (Fermentas, St Leon-Rot, Germany) together with 20 U of a ribonuclease inhibitor (RiboLock, Fermentas) as instructed in the manufacturer's protocol. cDNA was checked spectrophotometrically (Nanotrop) and by electrophoresis on a 1.5% agarose gel. Reproducibility was ensured by two independent repeats of the whole procedure.
Primer design using cDNA and vectorette PCR
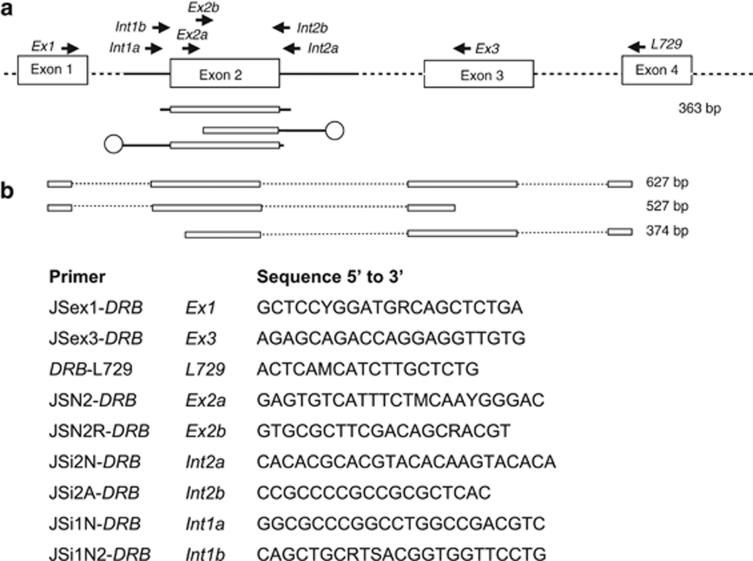
An MHC class II DRB cDNA alignment of different mammalian species was constructed by a BLAST search (http://www.ncbi.nlm.nih.gov) of the GenBank database. Primers complementary to conserved parts of exon 1 (Ex1) and exon 3 (Ex3) were designed (Figure 2). They were used together with primer L729 situated in exon 4 (Bowen et al., 2004, Figure 2) in different combinations to amplify DRB sequences from the cDNA of N. albiventris. PCR amplification was carried out in a total reaction volume of 20 μl, including 20 ng cDNA, 1 U of proofreading polymerase (HotStar HiFidelity Polymerase, Qiagen), 1 × HotStar HiFidelity PCR buffer, 0.3 m deoxynucleotid triphosphates (dNTPs) and 0.4 μ of each primer (Sigma-Aldrich, Steinheim, Germany). Thermocycling started with an initial denaturation step at 96 °C for 10 min, followed by 35 cycles of denaturation at 96 °C for 45 s, annealing at 58 °C for 45 s and elongation at 72 °C for 2 min. An additional 15 min elongation step followed the last cycle. PCR products were purified (Cycle pure Kit, Peqlab) and cloned using the pCR4-TOPO TA cloning Kit (Invitrogen). At least 23 recombinant clones per primer combination were amplified using the vector primers T7for and M13rev, sequenced using BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and analysed on ABI PRISM 310 Automated Genetic Analyzer (Applied Biosystems). All primer combinations amplified the same two MHC class II DRB alleles in N. albiventris. These Noal sequences were used to design the species-specific primers Ex2a and Ex2b (Figure 2) binding to conserved sites of exon 2 (inferred from the established cDNA alignment and available sequences of the bat Saccopteryx bilineata (GenBank accession numbers: EF533888-EF533900)), which were used for the subsequent first vectorette PCR assay.
Figure 2.
Positions and sequences of PCR primers to amplify the indicated fragments of the MHC class II DRB gene in N. albiventris based on gDNA (a) and cDNA (b). Boxes symbolise exons, dark lines mark introns and dotted lines mark not sequenced parts of the introns. Open circles indicate positions of ligated vectorettes (see Materials and Methods for details). The primer DRB-L729 is taken from Bowen et al. (2004).
Vectorette PCR is a method to amplify DNA fragments of interest, where the sequence information is only available on one side, and is described in detail by Ko et al. (2003). We constructed vectorette libraries with gDNA of five individuals. Up to 5 μg of gDNA, because of available template of each animal, were digested with restriction enzymes EcoRI, XapI, Fsp, and CspI (Fast Digest, Fermentas). In total, 1–5 μg gDNA were digested by 1–5 U restriction enzyme in a total volume of 50 or 100 μl depending on the amount of gDNA. Double-stranded vectorettes (1 μ) consisting of vect53 and vect57TTAA (EcoRI and XapI) or vect53 and vect57AT (Fsp and CspI) (Ko et al., 2003) were ligated to the sticky ends of the digested gDNA using 2 U of T4DNA ligase per 1 μg digested gDNA (Rapid DNA Ligation Kit, Fermentas).
Vectorette PCR assays consisted of two PCRs. The first PCR was followed by a second nested PCR to avoid false positives. A step-down scheme was always applied, using HotStar Taq Master Mix (Qiagen) according to the user's manual in a total reaction volume of 25 μl with the following conditions: initial activation of HotStar Taq and denaturation at 95 °C for 14 min, followed by 5 cycles of denaturation at 95 °C for 60 s, annealing at 67 °C for 60 s and elongation at 72 °C for 2 min, five cycles with annealing at 63 °C, followed by 15 cycles denaturation at 95 °C for 45 s, annealing at 60 °C for 45 s and extension at 72 °C for 120 s, 15 cycles with annealing at 57 °C and a final extension at 72 °C for 10 min. Amplification products were checked on a 1.5% agarose gel and purified with cycle Pure Kit (Peqlab).
The first vectorette PCR assay was performed with the primer Ex2a and the vectorette primer C20 (Ko et al., 2003), and the nested vectorette PCR included the nested primer Ex2b in combination with the nested vectorette primer B21 (Ko et al., 2003). The resulting intron 2 sequences allowed the design of Noctilio-specific intron 2 primers (Int2a and Int2b) (Figure 2). These were used to amplify intron 1 sequences in another vectorette PCR assay. Finally, according to the intron 1 sequences, the primers Int1a and Int1b were developed. Int1a and Int2a were used to amplify the whole 270 bp MHC DRB class II exon 2 and partial introns (45 bp in intron1, 48 bp in intron2, Figure 2) and were applied to genotype all 215 N. albiventris individuals.
Genotyping and allele identification
Amplification was performed in 25 μl volumes containing 0.4 μ of each primer (Sigma-Aldrich), 1 × reaction buffer (10 m Tris HCl, 50 m KCl, 0.1% Triton x-100, 0.2 mg ml−1 bovine serum albumin, MP Biomedicals, Heidelberg, Germany). 0.2 m of each dNTP and 1 U Taq polymerase (MP Biomedicals). Step-down PCR was performed as follows: initial incubation at 95 °C for 5 min, followed by 5 cycles of denaturation at 95 °C for 10 s, annealing at 67 °C for 10 s and elongation at 72 °C for 30 s, five cycles with annealing at 64 °C and 25 cycles denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s and elongation at 72 °C for 30 s, final extension was at 72 °C for 3 min. Amplicons were genotyped by single strand confirmation polymorphism on a polyacrylamide gel as described in Schad et al. (2004). Allele identification was done by excising the single strands from the gel matrix and diluting them in distilled water. Thereafter a reamplification of diluted single strands with primers Int1a and Int2a was performed before sequence analyses as described above. An autonomous amplicon with primer Int1b and Int2b of each individual was directly sequenced to confirm the individual single strand confirmation polymorphism pattern.
Statistical analyses
We edited and aligned nucleotide sequences manually using M 4.0 (Tamura et al., 2007). We also used this programme for calculating the P-distance of amino-acid sequences as a measurement of functional MHC class II DRB divergence (Nei and Kumar, 2000) and for analysing the relative rates of non-synonymous (dN) and synonymous (dS) base pair substitutions according to Nei and Gojobori (1986), applying the Jukes–Cantor correction for multiple hits (Jukes and Cantor, 1969). Calculations were applied for all sites and separately for putative ABS and non-ABS, assuming functional congruence to human ABS of the HLA-DR1 (DRA/DRB1*0101) molecule (Brown et al., 1993). The dN/dS ratios of all sites as well as for ABS and non-ABS separately were compared with an implemented Z-test (Nei and Kumar, 2000) to test for positive selection.
We identified species-specific positively selected sites (PSS) with maximum-likelihood analysis using C (included in P version 3.15 software package; Yang, 1997; Yang, 2007) and compared these PSS with the human ABS. First, we fitted models with different assumptions of selection patterns to the sequence data. We used the models M7 (β) and M8 (β and ω) as described in Yang et al. (2007). M7 served as a null model, where the ω-ratio varies according to the β- distribution and does not allow positive selected sites (0<ω<1). M8 adds a class of sites to account for the possible occurrence of PSS (ω>1). The models were compared using a likelihood ratio test by calculating the likelihood difference 2Δl=2(l1−l0) and then compared with a χ2-distribution, with the degrees of freedom equal to the difference in the number of estimated parameters (Yang and Bielawski, 2000). In the next step, after likelihood ratio test provided evidence for positive selection, the Bayes empirical Bayes method (Yang et al., 2005) integrated in C was used to identify the sites under positive selection with the cutoff posterior probability set at Pb=95%.
Allele frequencies, observed and expected heterozygosity, and deviation from Hardy–Weinberg expectations were estimated using the software A version 3.0 (Excoffier et al., 2005). Pairwise FST based on haplotype frequencies were calculated to infer population subdivision (10 000 permutations, Wright, 1951, 1965). Allelic richness (R) as a measure of the number of alleles independent of sample size was estimated using the rarefaction method as implemented in version (Goudet, 2001).
The χ2-tests were used to compare the number of heterozygote individuals between the sexes. Differences in the mean individual amino-acid distance of males and females were investigated by analysis of variance (ANOVA). Calculations were always two-tailed with significance level at α=0.05 and performed using S version 16.0 (SPSS Inc., Chicago, IL, USA). Bonferroni corrected significant levels were applied for multiple comparisons (Rice, 1989).
Results
Capture and sampling
In total, we caught 215 individuals (91 males and 123 females), 185 in Gamboa (59 males and 108 females) and 29 in BCI (22 males and 7 females). All of them were genotyped for the MHC class II DRB gene. In Gamboa, we caught bats from seven roosts. We included only colonies with more than 10 individuals in statistical analyses (colony A: N=52, 22 males and 30 females; colony B: N=74, 20 males and 54 females; colony C: N=27, 13 males and 14 females; and colony D: N=14, 4 males and 10 females).
MHC class II DRB diversity
Using cDNA of one male N. albiventris, all primer combinations amplified the same two MHC class II DRB alleles differing only in fragment length (628, 527 and 376 bp, respectively, Figure 2). We designed species-specific intron primers (Int1a and Int2a) for amplification of the whole MHC class II DRB exon 2 (270 bp) of N. albiventris flanked by short intron sequences with a total length of 363 bp (Figure 2) by means of the vectorette PCR approach based on gDNA. No indels or stop codons were found and no more than two alleles per individual were amplified, suggesting that a single DRB locus was expressed. We named this locus MhcNoal-DRB according to the established MHC nomenclature (Klein et al., 1990; Ellis et al., 2006). The nucleotide and deduced amino-acid sequence showed high homology with human (84%), canine (83%), equine (83%) and other mammalian class II DRB genes.
We detected 18 different alleles of Noal-DRB in the 215 individuals of N. albiventris (GenBank accession numbers: HM347941-HM347958). In the nucleotide sequences, we observed 71 (26.3%) variable positions and the alleles differed by 2 to 40 (average 26.3±2.9) nucleotide positions. All alleles had a unique amino-acid sequence, whereas 38 (43%) out of 89 amino acids were polymorphic. They differed by 1 to 26 (average 17.6±2.5) amino-acid positions.
The most common allele Noal-DRB*10 occurred at a frequency of 0.255 followed by Noal-DRB*02 (0.175) and Noal-DRB*04 (0.117). The remaining alleles occurred in 2 to 36 individuals with a frequency ranging between 0.005 and 0.089 (Table 1). Observed heterozygosity (0.902) was higher than expected (0.871) and did not deviate from Hardy–Weinberg expectations (Table 2). The individual Noal-DRB exon 2 distance of an individual based on amino-acid sequence ranged from 0 (homozygote) to 0.287 with an average of 0.177±0.078 (Table 2).
Table 1. Allele frequencies of MHC class II DRB alleles in the whole population and in subpopulations, roosting colonies and sexes.
| Level | All |
Subpopulations |
Roosting colonies |
Sexes |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Category | BCI | Gamboa | A | B | C | D | Females | Males | |
| N | 215 | 29 | 185 | 52 | 74 | 27 | 14 | 123 | 91 |
| NoalDRB*01 | 0.040 | 0.069 | 0.035 | 0.048 | 0.027 | 0.056 | 0.036 | 0.024 | 0.060 |
| NoalDRB*02 | 0.175 | 0.172 | 0.176 | 0.154 | 0.182 | 0.296 | 0.072 | 0.175 | 0.176 |
| NoalDRB*03 | 0.009 | — | 0.012 | 0.010 | 0.014 | — | — | 0.012 | 0.006 |
| NoalDRB*04 | 0.117 | 0.138 | 0.113 | 0.135 | 0.095 | 0.056 | 0.143 | 0.106 | 0.124 |
| NoalDRB*05 | 0.044 | 0.017 | 0.049 | 0.087 | 0.054 | 0.019 | — | 0.045 | 0.044 |
| NoalDRB*06 | 0.040 | 0.069 | 0.035 | 0.039 | 0.020 | 0.074 | 0.072 | 0.049 | 0.026 |
| NoalDRB*07 | 0.005 | — | 0.005 | 0.010 | 0.007 | — | — | 0.004 | 0.006 |
| NoalDRB*08 | 0.042 | 0.034 | 0.043 | 0.087 | 0.027 | 0.019 | 0.036 | 0.033 | 0.055 |
| NoalDRB*09 | 0.054 | 0.034 | 0.057 | 0.029 | 0.081 | 0.037 | 0.036 | 0.057 | 0.050 |
| NoalDRB*10 | 0.255 | 0.190 | 0.264 | 0.183 | 0.297 | 0.259 | 0.464 | 0.293 | 0.209 |
| NoalDRB*11 | 0.028 | 0.034 | 0.027 | 0.039 | 0.027 | — | — | 0.012 | 0.050 |
| NoalDRB*12 | 0.088 | 0.172 | 0.076 | 0.087 | 0.061 | 0.074 | 0.071 | 0.081 | 0.100 |
| NoalDRB*13 | 0.028 | — | 0.032 | 0.019 | 0.014 | 0.037 | 0.071 | 0.029 | 0.022 |
| NoalDRB*14 | 0.005 | 0.017 | 0.003 | — | 0.007 | — | — | 0.004 | 0.006 |
| NoalDRB*15 | 0.005 | — | 0.008 | — | 0.007 | 0.019 | — | 0.008 | - |
| NoalDRB*16 | 0.007 | — | 0.007 | 0.010 | 0.014 | 0.019 | — | 0.008 | 0.011 |
| NoalDRB*17 | 0.044 | 0.017 | 0.049 | 0.058 | 0.054 | 0.037 | — | 0.053 | 0.033 |
| NoalDRB*18 | 0.014 | 0.034 | 0.012 | 0.010 | 0.013 | — | — | 0.008 | 0.022 |
Abbreviations: BCI, Barro Colorado Island; MHC, major histocompatibility complex; N, sample size.
Table 2. MHC class II DRB exon 2 variability in N. albiventris in the whole population and in subpopulations, roosting colonies and sexes.
| Level | N | Number of alleles | R | Hobs | Hexp | Individual distance |
|---|---|---|---|---|---|---|
| All | 215 | 18 | 0.902 | 0.871 | 0.177±0.005 | |
| Category | ||||||
| Subpopulation | ||||||
| BCI | 29 | 13 | 13 | 0.931 | 0.886 | 0.189±0.013 |
| Gamboa | 185 | 18 | 13.6 | 0.817 | 0.868 | 0.175±0.006 |
| Roosting colonies | ||||||
| A | 52 | 16 | 11.1 | 0.981 | 0.901 | 0.190±0.009 |
| B | 74 | 18 | 10.4 | 0.838 | 0.856 | 0.164±0.010 |
| C | 27 | 13 | 10.1 | 0.889 | 0.839 | 0.166±0.016 |
| D | 14 | 9 | 9.0 | 0.857 | 0.767 | 0.171±0.022 |
| Sexes | ||||||
| Females | 123 | 18 | 17.2 | 0.854 | 0.856 | 0.164±0.008 |
| Males | 93 | 17 | 17.0 | 0.967 | 0.888 | 0.195±0.006 |
Abbreviations: BCI, Barro Colorado Island; Hexp, expected heterozygosity; Hobs, observed heterozygosity; Individual distance (±s.e.), individual amino-acid distance between alleles; N, sample size; R, allelic richness adjusted to the smallest sample size per level.
Evidence for historical selection
Two approaches were used to test for historical positive selection acting on the examined exon 2 of Noal-DRB locus. First, the averaged rates of non-synonymous (dN) and synonymous (dS) base pair substitutions of all sequences were calculated for all sites and separately for putative ABS and non-ABS, assuming functional congruence to human ABS of the HLA-DR1 molecule (Brown et al., 1993). Non-synonymous substitutions occurred at a significantly higher rate than synonymous ones (dN/dS=2.90, Z-test, P=0.004) especially in the regions that code for ABS (dN/dS=3.14, Z-test, P=0.002; Table 3). Second, we observed a significantly higher log likelihood estimate (2Δl=59.67, d.f.=2, P<0.0001) for model M8 (positive selection) than for its corresponding null model M7 (no positive selection). The Bayes empirical Bayes approach under model M8 inferred 17 significant sites (PSS) to be under positive selection with the cutoff posterior probability set at 95%. In total, 14 of these sites were congruent with predicted ABS of the human HLA-DR1 β-chain (Brown et al., 1993). The other three sites were located in close proximity within one to two amino-acid positions to the human ABS (Table 4, Figure 3). In the Noal-DRB sequences, all but one (61 W) of certain amino-acid positions, which are conserved in human HLA-DR1 molecules (Brown et al., 1993; Stern et al., 1994), were also conserved presenting the identical amino acids. At position 61, tryptophan was replaced by leucine in six Noal alleles (33.3%) and identified as PSS (Table 4, Figure 3).
Table 3. Non-synonymous (dN) and synonymous (dS) substitutions (±s.e.) as well as their ratio in ABS and non-ABS assuming concordance with the human HLA-DR1 molecule (Brown et al., 1993).
| Region | N | dN | dS | dN/dS | P |
|---|---|---|---|---|---|
| ABS | 25 | 0.353±0.077 | 0.112±0.044 | 3.139 | 0.002 |
| Non-ABS | 64 | 0.053±0.013 | 0.034±0.018 | 0.942 | NS |
| All | 89 | 0.124±0.021 | 0.055±0.018 | 2.904 | 0.004 |
Abbreviations: ABS, antigen-binding sites; N, number of codons in each category; NS, not significant; P, probability (α⩽0.05) that dN and dS are different using a Z-test.
Table 4. Identification of species-specific PSS by likelihood analysis in MHC class II DRB exon 2 amino-acid sequences of N. albiventris.
| PSS | Mean ω | Pb (ω>1) | Distance to ABS |
|---|---|---|---|
| 9 | 4.561±0.496 | 1.000** | 0 |
| 11 | 4.561±0.496 | 1.000** | 0 |
| 12 | 4.490±0.693 | 0.982* | 0 |
| 13 | 4.556±0.514 | 0.999** | 0 |
| 16 | 3.908±1.434 | 0.836 | 2 |
| 18 | 4.353±0.950 | 0.948 | 4 |
| 28 | 4.464±0.740 | 0.976* | 0 |
| 34 | 3.633±1.604 | 0.768 | 4 |
| 37 | 4.552±0.526 | 0.998** | 0 |
| 47 | 3.460±1.677 | 0.724 | 0 |
| 57 | 4.561±0.496 | 1.000** | 0 |
| 60 | 3.733±1.530 | 0.793 | 1 |
| 61 | 4.512±0.643 | 0.988* | 1 |
| 63 | 4.462±0.747 | 0.975* | 0 |
| 66 | 4.334±0.970 | 0.944 | 0 |
| 67 | 4.524±0.604 | 0.991** | 0 |
| 70 | 4.560±0.493 | 1.000** | 0 |
| 71 | 4.561±0.496 | 1.000** | 0 |
| 74 | 4.561±0.496 | 1.000** | 0 |
| 76 | 3.581±1.650 | 0.755 | 2 |
| 77 | 4.529±0.592 | 0.992** | 1 |
| 78 | 4.559±0.503 | 1.000** | 0 |
| 86 | 4.561±0.496 | 1.000** | 0 |
| 88 | 3.646±1.731 | 0.778 | 0 |
| 92 | 4.545±0.549 | 0.996** | 2 |
Abbreviations: ABS, antigen-binding sites; Distance to ABS, amino acids to nearest human ABS of the HLA-DR1 β-chain (Brown et al., 1993); Mean ω (±s.e.), ratio of non-synonymous and synonymous substitutions at PSS and its probability (Pb (ω>1)) using a cutoff posterior probability of 95% PSS, positively selected sites, numbers indicate the amino-acid positions in the β1-domain.
**α⩽0.01, *α⩽0.05.
Figure 3.
Amino-acid sequence variation of 18 MHC class II DRB exon 2 alleles of N. albiventris and the human HLA-DRB1*0101 sequence. Dots mark identity with the top sequence. Numbers indicate the amino-acid positions of the β1-domain, * signify the amino-acid positions of ABS and conserved sites (ConsSites) of the human HLA-DR1 β-chain (Brown et al., 1993; Stern et al., 1994). PSS indicates species-specific positive selected sites identified by C (Yang et al., 2005).
Population structure and MHC
We observed no genetic differentiation between the two subpopulations of Gamboa and BCI (FST=0.003, P=0.23). Allele frequencies of the two subpopulations are shown in Table 1. Numbers of alleles were similar after correction for differences in sample size. Observed heterozygosity was high in both subpopulations without a significant deviation from Hardy–Weinberg expectations (Table 2).
All four roosting colonies with more than 10 individuals captured in the village of Gamboa showed high levels of heterozygosity and allelic richness (R) ranging from 9 to 11.1. Observed heterozygosity exceeded the expected value in all but one colony (colony B) and did not deviate significantly from Hardy–Weinberg equilibrium (Table 2). Pairwise FST statistics showed a slight differentiation between colonies A, C and D but this significance was lost after Bonferroni correction (α′ ⩽0.008; Table 5).
Table 5. Pairwise differentiation between roosting colonies using conventional F-statistic based on haplotype frequencies (Wright, 1965).
| Colony | A | B | C | D |
|---|---|---|---|---|
| A | NS | NS | 0.010 | |
| B | 0.006 | NS | NS | |
| C | 0.012 | 0.002 | 0.031 | |
| D | 0.036 | 0.013 | 0.036 |
Abbreviation: NS, not significant.
FST values are provided below the diagonal and corresponding P-values above diagonal. Bonferroni corrected significance level α′⩽0.008.
We found no significant differentiation within males and females when comparing colonies (males: range of FST: 0.001–0.026, P-values not significant; females: FST: <0.001–0.039, P-values not significant; Bonferroni corrected significance level α′⩽0.008). We found no significant differentiation between males and females within colonies either (FSTA=0.009, P=0.87; FSTB=0.001, P=0.44; FSTC=0.007, P=0.54; FSTD=0.030, P=0.82; and FSTAll=0.002, P=0.87).
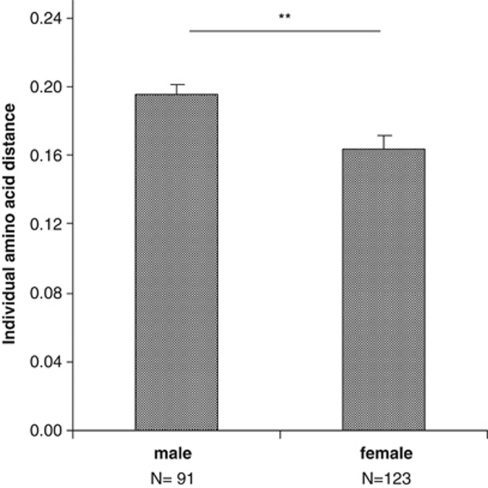
However, males and females differed significantly in their individual amino-acid distance (ANOVA: F=8.48, P=0.004, d.f.=1, Figure 4, Table 2). Furthermore, the observed heterozygosity of males was significantly higher than in females (overall: χ2=7.73, d.f.=1, P=0.005). The analyses were not significant when colonies were analysed separately (A: χ2=0.81, P=0.36; B: χ2=2.54, P=0.11; C: χ2=3.13, P=0.07; and D: χ2=0.93, P=0.33). In males, the observed heterozygosity exceeded the expected value in the overall sample as well as in all colonies analysed separately but did not deviate significantly from Hardy–Weinberg equilibrium (overall: Hobs=0.97, HexP=0.89; A: Hobs=1.00, HexP=0.91; B: Hobs=0.96, HexP=0.89; C: Hobs=1.00, HexP=0.88; D: Hobs=1.00, HexP=0.89; and BCI: Hobs=1.00, HexP=0.89). In females, the observed heterozygosity was always lower than in males but close to the expected value in the overall sample as well as in two out of four colonies (overall: Hobs=0.85, HexP=0.86; A: Hobs=0.96, HexP=0.90; B: Hobs=0.83, HexP=0.86; C: Hobs=0.78, HexP=0.79; D: Hobs=0.75, HexP=0.75; and BCI: Hobs=1.00, HexP=0.92; Table 2).
Figure 4.
Mean (±s.e.) amino-acid distance between individual MHC class II DRB exon 2 alleles of N. albiventris in males and females. ANOVA: F=8.48, d.f.=1, **P=0.004; N, sample size.
Discussion
To date, not much is known about the genetic structure and polymorphism of the MHC in bats (Meyer and Brunner, 2007) mainly because of a lack of sequence data for primer design. This is astonishing given the size of the order, the importance of olfactory signals for social communication in bats and their zoonotic relevance as reservoir hosts for many pathogens. Therefore, our primary aim was to determine MHC class II DRB exon 2 diversity in the lesser bulldog bat, N. albiventris. Further, we tested for evidence of positive selection acting above the evolutionary history of the species. And third, we were interested whether local adaptation and sexual selection may shape the contemporary genetic structure of N. albiventris in a Central American population.
MHC class II DRB diversity
Well-designed primers are essential for population genetic analyses. In this study, cDNA analyses followed by a vectorette PCR approach offered the possibility of intron mapping. Species-specific primers binding to conserved segments of the flanking introns of N. albiventris were designed to amplify the whole DRB exon 2. The reliability of the resulting species-specific sequence patterns of the locus of interest is very high because the incidence of non-amplifying alleles can be neglected. This might turn into a problem when exon primers are used to amplify parts of polymorphic genes. The occurrence of non-amplifying alleles would preclude the use of data for many population genetic purposes because they can seriously bias population genetic analyses (Dakin and Avise, 2004; Cummings et al., 2010).
We found evidence for a single expressed MHC class II DRB locus in N. albiventris with moderate allelic variability of 18 alleles detected in 215 individuals. This allelic variability is within the range of MHC class II DRB polymorphism of other mammalian species (for example, Sommer, 2005). The only other DRB gene studied in a bat, Saccopteryx bilineata, revealed a rather low allelic variability (11 alleles in 85 individuals; Meyer and Brunner, 2007). However, this might be an underestimate of the actual variability because preliminary studies on RNA and DNA with species-specific developed primers revealed evidence for at least five DRB loci in Saccopteryx bilineata (Schad et al., unpublished data). In general, the occurrence of different MHC class II genes (DP, DO, DM, DQ and DR) is conserved in mammals. However, the number of functional α- and β-genes is highly variable because of species-specific local duplication events. Yet, it can even vary between individuals of the same species (see Kumanovics et al., 2003). The DRB is the most widely studied and usually the most diverse class II gene, not only with respect to high allelic richness but also in terms of gene duplications (for example, Doxiadis et al., 2000; Bowen et al., 2004; Babik et al., 2005; Schwensow et al., 2007). We cannot rule out that other MHC class II genes may provide a higher variability in bats as it is described, for instance, in the cetacean MHC class II DQB gene (Baker et al., 2006). As a future task, it will be necessary to investigate other loci to evaluate MHC diversity in bats more comprehensively.
Evidence for historical selection
We found a higher rate of non-synonymous versus synonymous nucleotide substitutions over the entire sequence and especially in the putative ABS but not in non-ABS (Brown et al., 1993). Such elevated dN/dS ratios are a widely accepted sign for historical positive selection in polymorphic MHC genes (Hughes and Nei 1988, 1989; Nei and Kumar, 2000; Nielsen, 2001; Hughes, 2007). They indicate positive selection on ABS acting above the evolutionary history of the species (Hedrick, 1999; Bernatchez and Landry, 2003; Sommer, 2005). We subsequently confirmed these results in a species-specific analysis where different codon evolution models were compared by maximum-likelihood analyses (Yang and Bielawski, 2000; Yang, 2007). This method has been suggested as a powerful tool with a high likelihood of detecting effects of positive selection (Wong et al., 2004). Our data fitted best with the model incorporating positive selection and 17 species-specific PSS were identified by the Bayes empirical Bayes analysis. A total of 14 were congruent with human ABS of the HLA-DR1 β-chain. Three PSS were situated outside the human ABS and some of the human ABS were not identified as PSS in Noal-DRB. Comparable results have been reported from other species (Kundu and Faulkes, 2004; Schwensow et al., 2007; Babik et al., 2008; Meyer-Lucht et al., 2008). In general, high congruence of PSS with human ABS is assumed to demonstrate homologous functionality of the molecule. Contrarily, human ABS that are not identified as PSS might be not involved in the antigen recognition and binding in the respective species. Furthermore, in most species investigated so far additional PSS have been reported. These findings suggest species-specific selection pressures acting on MHC genes because of a different pathogen exposure. In addition, certain amino-acid residuals of HLA-DR1 molecules are highly conserved and involved in universal hydrogen bond of antigens (61 W, 81 H and 82 N) or are responsible for the stability of DR1 heterodimers in building salt bridges between the dimers (52 E and 55 R) (Brown et al., 1993; Stern et al., 1994). Also in the Noal-DRB sequences, all of these positions, except position 61, were conserved indicating similar conserved functionality of the molecules.
Population structure and MHC
Mating behaviour as a correlate and driver of social structure has been suggested in addition to pathogens as another main subject of selection on MHC loci in natural populations (Hambuch and Lacey, 2002; Kundu and Faulkes, 2004; Cutrera and Lacey, 2006). Animals form social groups and colonies in response to cooperative interactions as well as mating tactics. Furthermore, patterns of genetic subdivision are also shaped by the extent and nature of philopatric behaviour (Travis et al., 1995; Sommer et al., 2002; Solomon, 2003; Cutrera and Lacey, 2006). We examined the genetic structure of three different levels of social formations or units (subpopulation, roosting colonies and sexes) in our population of N. albiventris. We found no genetic differentiation between the two subpopulations (Gamboa and BCI, separated by 15 km), indicating the presence of gene flow at a larger spatial scale. Both subpopulations showed similar levels of polymorphism at the Noal-DRB locus. Observed heterozygosity was high in the two subpopulations and the allele frequencies and the number of alleles were similar after correcting for differences in sample sizes. This implies an equivalent selection pressure maintaining diversity at the Noal-DRB locus in both subpopulations in the recent past.
To investigate social structure based on roosting habits, we compared four colonies in the village of Gamboa, all located in the range of 1.5 km2. All colonies showed similar levels of genetic variation (heterozygosity, allelic richness and allele frequencies). F-statistics revealed only limited effects of subdivisions. The colony D showed a slight differentiation compared with the colonies A and C (not significant after Bonferroni correction), which might be rather the result of missing rare alleles because of small sample size than an effect of population structure based on roosting habits. Dechmann et al. (2009) distinguished social groups of females (2–5 individuals) by the fact that they emerged simultaneously from a roost. We could not find genetic differences between male and female members of different colonies. The composition of colonies might vary in time, indicating a fission–fusion society rather than stable associations. Neutral markers would offer the possibility to gain insights into population dynamic processes like kin relationships of social groups of females and roosting colonies. Thus, further ecological studies on demographic structure as well as genetic analyses adding neutral markers will help to fully understand the social system of this species.
We tested for gender-specific differences in the MHC constitution to assess the occurrence of sexual selection. We found no population differentiation between males and females. However, in the overall sample, males showed a significantly higher heterozygosity rate and also a higher individual amino-acid distance than females. In males, the observed heterozygosity exceeded the expected value in the overall sample as well as in all within-colony comparisons. In females, the observed heterozygosities were always lower than in males but were almost identical with the expected ones in the overall sample and in two out of four colonies. As in both sexes observed and expected heterozygosities did not significantly deviate from Hardy–Weinberg expectations, it remains unclear whether the difference between sexes is because of a higher heterozygosity rate in males or a lower heterozygosity rate in females. An increased heterozygosity rate in males could indicate balancing selection in form of a heterozygote advantage and a deficit in heterozygote females could suggest a reduced selection pressure to maintain diversity in females. It might be that the selection intensity in the investigated Noctilio population is not strong enough to detect significant deviations from Hardy–Weinberg expectations. It is well known that levels of allelic diversity in relation to sample size have an effect on the statistical power to detect significant deviations from Hardy–Weinberg expectations (Seddon and Ellegren, 2004). We did not find small-scale population structure that would offer a likely explanation for this gender-specific difference. Male-based long-distance dispersal is unlikely as it would promote the occurrence of new alleles in the male population, raising the overall polymorphism at the MHC locus that we did not observe. A methodological error because of DNA quality differences between the sexes seems also to be unlikely because collection and treatment of samples have been the same throughout the study. Although direct female choice for heterozygote males would lead to heterozygote offspring in general including daughters, it cannot explain the sex-specific bias in heterozygosity. Currently, the differences in heterozygosity in males and females is most likely due to MHC-mediated postcopulatory mechanisms (for example, caused by myotic drive, gametic selection and maternal–fetal interactions) or by sex-specific survival differences that result in an increased rate of MHC heterozygote males or a deficit in heterozygote females.
The possibility of the existence of postcopulatory mechanisms resulting in sex-specific differences in MHC heterozygosity was reported only in a few studies so far. Dorak et al. (2002) found an increased heterozygosity for MHC class II DRB lineages in newborn male babies and suggested that negative selection of homozygotes might be restricted to male offspring only. A deficit in MHC homozygosity in newborn males was also observed in mice (Hamilton and Hellstrom 1978) and rats (Palm 1969, 1970, 1974). Some studies in humans investigated the compatibility at different HLA loci between mothers and infants and observed differences in the sex ratio assuming a different fetal loss in males and females. The results are heterogeneous and differ between HLA loci (Ober et al., 1987; Astolfi et al., 1990, 1996). In all of these studies, the underlying mechanisms have not been investigated. The ongoing discussion has been reviewed by Fernandez et al. (1999) and Ziegler et al. (2005). The overall conclusion is that the MHC is critical for numerous aspects of mammalian reproduction concerning spermatogenesis (Ziegler et al., 2002, 2005), a sperm-selective egg-cumulus complex (Wedekind et al., 1996; Rülicke et al., 1998; Eisenach and Giojalas, 2006), and viability and development of the foetus (Gill, 1992; Ober et al., 1987; Astolfi et al., 1990; Wedekind et al., 1996; Ziegler et al., 2005). However, in all these processes the selective forces might act because of MHC-linked genes, for example, olfactory receptor genes, transcription factors and others rather than by the MHC itself (Ho et al., 1990; Gill, 1992; Ziegler et al., 2002, 2005; Eisenach and Giojalas, 2006). To the best of our knowledge, sex-specific differences in offspring survival based on MHC heterozygosity have not been reported yet. But they have been investigated using a microsatellite-based measure of outbreeding (mean d2) in a few species. In the bat Rhinolophus ferrumequinum, outbreeding was positively associated with significantly increased survival in male offspring only (Rossiter et al., 2001). The authors postulate that outbreeding at the microsatellite markers reflects immunocompetence, which in turn influences mortality. They also suggest that characterisation of MHC loci may provide a suitable test for their hypothesis. Even so, in the red deer (Cervus elaphus) a contrary pattern was found as male offspring survival was negatively associated with outbreeding at microsatellite markers (Coulson et al., 1999). In mice, a reduced survivorship was reported for inbred adult males but not for females, most likely as a consequence of aggressive interactions of males in the defence of territories (Meagher et al., 2000). Ongoing studies might reveal the mechanisms that have contributed to the sex-biased diversity pattern in the investigated population of N. albiventris. In addition, neutral markers would provide more detailed information about social structure, dispersal behaviour and gene flow of both sexes.
In this first study on the MHC class II variability of the lesser bulldog bat, we detected high genetic variation and evidence for historical positive selection acting on a single expressed Noal-DRB locus. The polymorphism at the antigen-binding region of the molecule is considered as the precondition to cope with a variety of pathogens. No population differentiation between subpopulations, roosting colonies and sexes was observed, but males revealed a significantly higher heterozygosity rate and genetic variability in terms of the genetic distance between the individual MHC alleles than females. We are aware that at this state of the investigation no conclusions on the underlying mechanisms can be made but our data can provide the basis for further research on the role of the MHC constitution in host–pathogen interactions, individual body odours and sexual selection in a highly interesting bat species.
Acknowledgments
We thank Silke L Voigt-Heucke, Antje Kretzschmar, Megan Lupek, Martin Wikelski and Apanie Wood for help with catching bats and sample collection. We thank Wieslaw Babik for initial advice on the vectorette PCR approach. We are grateful to Charles Gunn for linguistic advice. We thank three anonymous reviewers for helpful comments on former versions of the manuscript. The German Science Foundation funded C. Voigt, S. Speck and D. Dechmann (VO 890/11-1). All capture and handling of animals, as well as collection and export of samples were done in concordance with Panamanian laws under permits from the Panamanian authorities (ANAM) and under an IUCAC protocol. We are indebted to the Smithsonian Tropical Research Institute and their staff for help with logistics and interaction with the Panamanian authorities.
The authors declare no conflict of interest.
References
- Altringham JD, Fenton MB.2003Sensory ecology and communication in the chiropteraIn: Kunz TH, Fenton MB (eds).Bat Ecology University of Chicago Press: Chicago; 90–118. [Google Scholar]
- Astolfi P, Cuccia M, Caruso C, Favoino B, Fazzari M, Mantovani V, et al. Sharing at the major histocompatibility complex affects secondary sex ratio in differing ways. Hum Hered. 1996;46:155–165. doi: 10.1159/000154345. [DOI] [PubMed] [Google Scholar]
- Astolfi P, Martinetti M, Gigli-Berzolari F, Cuccia M. The effect of parental and maternal-fetal histocompatibility at MHC on sex ratio in offspring. Tissue Antigens. 1990;35:172–177. doi: 10.1111/j.1399-0039.1990.tb01775.x. [DOI] [PubMed] [Google Scholar]
- Babik W, Durka W, Radwan J. Sequence diversity of the MHC DRB gene in the Eurasian beaver (Castor fiber) Mol Ecol. 2005;14:4249–4257. doi: 10.1111/j.1365-294X.2005.02751.x. [DOI] [PubMed] [Google Scholar]
- Babik W, Pabijan M, Radwan J. Contrasting patterns of variation in MHC loci in the Alpine newt. Mol Ecol. 2008;17:2339–2355. doi: 10.1111/j.1365-294X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- Baker CS, Vant MD, Dalebout ML, Lento GM, O'Brien SJ, Yuhki N. Diversity and duplication of DQB and DRB-like genes of the MHC in baleen whales (suborder: Mysticeti) Immunogenetics. 2006;58:283–296. doi: 10.1007/s00251-006-0080-y. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years. J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Bouchard S. Sex discrimination and roostmate recognition by olfactory cues in the African bats, Mops condylurus and Chaerephon pumilus (Chiroptera: Molossidae) J Zool. 2001;254:109–117. [Google Scholar]
- Bowen L, Aldridge BM, Gulland F, Van Bonn W, DeLong R, Melin S, et al. Class II multiformity generated by variable MHC-DRB region configurations in the California sea lion (Zalophus californianus) Immunogenetics. 2004;56:12–27. doi: 10.1007/s00251-004-0655-4. [DOI] [PubMed] [Google Scholar]
- Brook AP, Decker DM. Lipid compounds in secretions of fishing Bat, Noctilio leporinus (Chiroptera: Noctilionidae) J Chem Ecol. 1996;22:1411–1428. doi: 10.1007/BF02027721. [DOI] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers B, Wibbelt G, Voigt CC. Sex specific differences of facial glands in the greater sac-winged bat Saccopteryxbilineata. Zoomorphology. 2009;128:37–43. [Google Scholar]
- Chaix R, Cao C, Donnelly P. Is mate choice in humans MHC-dependent. PloSGenet. 2008;4:e1000184. doi: 10.1371/journal.pgen.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesser RK. Gene diversity and female philopatry. Genetics. 1990;127:437–447. doi: 10.1093/genetics/127.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson TN, Albon SD, Slate J, Pemberton JM. Microsatellite loci reveal sex-dependent responses to inbreeding and outbreeding in red deer calves. Evolution. 1999;53:1951–1960. doi: 10.1111/j.1558-5646.1999.tb04575.x. [DOI] [PubMed] [Google Scholar]
- Cummings SM, McMullen M, Joyce DA, van Osterhout C. Solutions for PCR, cloning and sequencing errors in population genetic analyes. Conserv Genet. 2010;11:1095–1097. [Google Scholar]
- Cutrera AP, Lacey EA. Major histocompatibility complex variation in talas tuco-tucos: the influence of demography on selection. J Mammal. 2006;87:706–716. [Google Scholar]
- Dakin EE, Avise JC. Microsatellite null alleles in parentage analysis. Heredity. 2004;93:504–509. doi: 10.1038/sj.hdy.6800545. [DOI] [PubMed] [Google Scholar]
- Dapson RW, Studier EH, Buckingham MJ, Studier AL. Histochemistry of odoriferous secretions from integumentary glands in three species of bats. J Mammal. 1977;58:531–535. [PubMed] [Google Scholar]
- Dechmann DKN, Heucke S, Giuggioli L, Safi K, Voigt CC, Wikelski M. Experimental evidence for group hunting via eavesdropping in echolocating bats. Proc R Soc B. 2009;276:2721–2728. doi: 10.1098/rspb.2009.0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- Dorak MT, Lawson T, Machulla HKG, Mills KI, Burnett AK. Increased heterozygosity for MHC class II lineages in newborn males. Genes Immun. 2002;3:263–269. doi: 10.1038/sj.gene.6363862. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Ottig N, de Groot NG, Noort R, Bontrop RE. Unprecedented polymorphism of Mhc-DRB region configurations in Rhesus macaques. J Immunol. 2000;164:3193–3199. doi: 10.4049/jimmunol.164.6.3193. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Potts WK.1996Polymorphism of genes in the major histocompatibility complex (MHC): implications for conservation geneticsIn: Smith TB, Wayne RK (eds).Molecular Genetic Approaches in Conservation Oxford University Press: New York; 214–237. [Google Scholar]
- Ellis SA, Bontrop RE, Antczak DF, Ballingall K, Davies CJ, Kaufman J, et al. ISAG/IUIS-VIC comparative MHC nomenclature committee report, 2005. Immunogenetics. 2006;57:953–958. doi: 10.1007/s00251-005-0071-4. [DOI] [PubMed] [Google Scholar]
- Eisenach M, Giojalas LC. Sperm guidance in mammals—an unpaved road to the egg. Mol Cell Biol. 2006;7:276–285. doi: 10.1038/nrm1893. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fernandez N, Cooper J, Spinks M, AbdElrahman M, Fiszer D, Kurpisz M, et al. A critical review of the role of the major histocompatibility complex in fertilisation, preimplantation development and feto-maternal interactions. Human Reprod Update. 1999;5:234–248. doi: 10.1093/humupd/5.3.234. [DOI] [PubMed] [Google Scholar]
- Field HE. Bats and emerging zoonoses: henipaviruses and SARS. Zoonoses Public Hlth. 2009;56 (6–7:278–284. doi: 10.1111/j.1863-2378.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- Gill TJ., III Invited editorial: influence of MHC and MHC-linked genes on reproduction. Am J Hum Genet. 1992;50:1–5. [PMC free article] [PubMed] [Google Scholar]
- Goudet2001FSTAT version, a program to estimate and test gene diversities and fixationAvailable from http://www.unil.ch/izea/softwares/fstat.html .
- Gustin MK, McCracken GF. Scent recognition in the Mexican free-tailed bat, Tadarida brasiliensis mexicana. Anim Behav. 1987;35:13–19. [Google Scholar]
- Hambuch TM, Lacey EA. Enhanced selection for MHC diversity in social tuco-tucos. Evolution. 2002;56:841–845. doi: 10.1111/j.0014-3820.2002.tb01395.x. [DOI] [PubMed] [Google Scholar]
- Hamilton MS, Hellstrom I. Selection for histocompatibility progeny in mice. Biol Reprod. 1978;19:267–270. doi: 10.1095/biolreprod19.2.267. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56:1902–1908. doi: 10.1111/j.0014-3820.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW, Black FL. HLA and mate selection: no evidence in South Amerindians. Am J Hum Genet. 1997;61:497–504. doi: 10.1086/515519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AVS.1991HLA associations with Malaria in Africa: some implications for MHC evolutionIn: Klein J, Klein D (eds).Molecular Evolution of the Major Histocompatibility Complex Springer: Berlin, Germany; 403–419. [Google Scholar]
- Ho HN, Gill TJ, III, Hseih KP, Hseih HJ, Yee TY. Sharing of human leukocyte antigens (HLA) in primary and secondary recurrent spontaneous abortions. Am J Obstet Gyneol. 1990;163:178–188. doi: 10.1016/s0002-9378(11)90696-6. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmonen P, Stundner G, Thoss M, Penn DJ. Females prefer the scent of outbred males: good-genes as heterozygosity. BMC Evol Biol. 2009;9:104. doi: 10.1186/1471-2148-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Travers P. Immunology. Spektrum Akademischer Verlag GmbH: Heidelberg, Berlin, Oxford; 2002. [Google Scholar]
- Jukes TH, Cantor CR.1969Evolution of protein moleculesIn: Munroe HN (ed).Mammalian Protein Metabolism Academic Press: New York; 21–132. [Google Scholar]
- Klein J. Natural History of the Major Histocompatibility Complex. Wiley & Sons: New York; 1986. [Google Scholar]
- Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, et al. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutrality theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Ko WY, David RM, Akashi H. Molecular phylogeny of the Drosophila melanogaster species subgroup. J Mol Evol. 2003;57:562–573. doi: 10.1007/s00239-003-2510-x. [DOI] [PubMed] [Google Scholar]
- Kumanovics A, Takada T, Fischer Lindahl K. Genomic organization of the mammalian MHC. Annu Rev Immunol. 2003;21:629–657. doi: 10.1146/annurev.immunol.21.090501.080116. [DOI] [PubMed] [Google Scholar]
- Kundu S, Faulkes CG. Patterns of MHC selection in African mole rats, family bathyergidae: the effects of sociality and habitat. Proc R Soc Lond B. 2004;271:273–278. doi: 10.1098/rspb.2003.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J, Opiekun MC, Matsumura K, Preti G, Yamazaki K, Beauchamp GK. Major histocompatibility complex-regulated odor types: peptide-free urinary volatile signals. Phys Behav. 2009;96:184–188. doi: 10.1016/j.physbeh.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Matocq MD, Lacy EA. Philopatry, kin clusters, and genetic relatedness in a population of woodrats (Neotoma macrotis) Behav Ecol. 2004;15:647–653. [Google Scholar]
- McCracken GF, Wilkinson GS.2000Bat mating systemsIn: Crichton EG, Krutzsch PH (eds).Reproductive biology of bats Academic Press: San Diego, CA; 321–362. [Google Scholar]
- Meagher S, Penn DJ, Potts WK. Male-male competition magnifies inbreeding depression in wild house mice. Proc Natl Am Soc USA. 2000;97:3324–3329. doi: 10.1073/pnas.060284797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lucht Y, Otten C, Püttker T, Sommer S. Selection, diversity and evolutionary patterns of MHC class II DAB in free ranging neotropical marsupials. BMC Genetics. 2008;9:39. doi: 10.1186/1471-2156-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinski M. The major histocompatibility complex, sexual selection, and mate choice. Ann Rev Ecol, Evol Syst. 2006;37:159–186. [Google Scholar]
- Meyer F, Brunner A. Non-neutral evolution of the major histocompatibility complex class II gene DRB1 in the sac-winged bat Saccopteryx bilineata. Heredity. 2007;99:257–264. doi: 10.1038/sj.hdy.6800989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press, New York; 2000. [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- Ober C, Simpson JL, Ward M, Radvany RM, Andersen R, Elias S, et al. Prenatal effects of maternal-fetal HLA compatibility. Am J Reprod Immunol Microbiol. 1987;15:141–149. [PubMed] [Google Scholar]
- Ober C, Van der Ven K.1997Immunogenetics of reproduction: an overviewIn: Olding LB (ed).Current Topics in Microbiology and Immunology. Springer-Verlag: Berlin; 1–23. [DOI] [PubMed] [Google Scholar]
- Palm J. Association of maternal genotype and excess heterozygosity for Ag-B histocompatibility antigens among male rats. Transplant Proc. 1969;1:82–84. [PubMed] [Google Scholar]
- Palm J. Maternal-fetal interactions and histocompatibility antigen polymorphisms. Transplant Proc. 1970;2:162–173. [PubMed] [Google Scholar]
- Palm J. Maternal-fetal histocompatibility in rats: an escape from adversity. Cancer Res. 1974;34:2061–2065. [PubMed] [Google Scholar]
- Parham P, Otha N. Population biology of antigen-presentation by MHC class I molecules. Science. 1996;272:67–79. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- Patterson S, Pemberton JM. No evidence for major histocompatibility complex-dependent mating patterns in a free-living ruminant population. Proc R Soc Lond B. 1997;264:1813–1819. doi: 10.1098/rspb.1997.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn DJ, Potts WK. How do major histocompatibility complex genes influence odor and mating preference. Adv Immunol. 1998;69:411–436. doi: 10.1016/s0065-2776(08)60612-4. [DOI] [PubMed] [Google Scholar]
- Penn DJ, Potts WK. The evolution of mating preferences and major histocompatibility complex genes. Am Nat. 1999;153:145–164. doi: 10.1086/303166. [DOI] [PubMed] [Google Scholar]
- Piertney SB, Oliver MK. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- Quay WB.1970Integument and derivativesIn: Wimsatt WA (ed).Biology of bats Academic Press: New York; 1–56. [Google Scholar]
- Rice WR. Analysing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rossiter SJ, Jones G, Ransome RD, Barratt EM. Outbreeding increases offspring survival in wild greater horseshoe bats (Rhinolophus ferrumequinum) Proc R Soc Lond B. 2001;268:1055–1061. doi: 10.1098/rspb.2001.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rülicke T, Chapuisat M, Homberger FR, Macas E, Wedekind C. MHC-genotype of progeny influenced by parental infection. Proc R Soc Lond B. 1998;265:711–716. doi: 10.1098/rspb.1998.0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi K, Kerth G. Secretions of the interaural gland contain information about individuality and colony membership in the Bechstein's bat. Anim Behav. 2003;65:363–369. [Google Scholar]
- Schad J, Sommer S, Ganzhorn J. MHC variability of a small lemur in the littoral forest fragments of southeastern Madagascar. Conserv Genet. 2004;5:299–309. [Google Scholar]
- Schwensow N, Fietz J, Dausmann KH, Sommer S. Neutral versus adaptive genetic variation in parasite resistance: importance of major histocompatibility complex supertypes in a free-ranging primate. Heredity. 2007;99:265–277. doi: 10.1038/sj.hdy.6800993. [DOI] [PubMed] [Google Scholar]
- Schwensow N, Fietz J, Dausmann K, Sommer S. MHC-associated mating strategies and the importance of overall genetic diversity in an obligate pair living primate. Evol Ecol. 2008;22:617–636. [Google Scholar]
- Scully WMR, Fenton MB, Saleuddin ASM. A histological examination of the holding sacs and glandular scent organs of some bat species (Emballonuridae, Hipposideridae, Phyllostomidae, Vespertilionidae and Molossidae) Can J Zool. 2000;78:613–623. [Google Scholar]
- Seddon JM, Ellegren H. A temporal analysis shows major histocompatibility complex loci in the Scandinavian wolf population are consistent with neutral evolution. Proc R Soc Lond B. 2004;271:2283–2291. doi: 10.1098/rspb.2004.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade RW, McCallum HI. Overdominant vs frequency-dependent selection at MHC loci. Genetics. 1992;132:861–862. doi: 10.1093/genetics/132.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon NG. A re-examination of factors influencing philopatry in rodents. J Mammal. 2003;84:1182–1197. [Google Scholar]
- Sommer S, Schwab D, Ganzhorn JU. MHC diversity of endemic Malagasy rodents in relation to geographic range and mating system. Behav Ecol Sociobiol. 2002;51:214–221. [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc R Soc Lond B. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern LJ, Brown JH, Jardetzky JCG, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- Studier EH, Lavoie KH. Microbial involvement in scent production in noctilionid bats. J Mammal. 1984;65:711–714. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis SE, Slobodchikoff CN, Keim P. Ecological and demographic effects on intraspecific variation in the social system of prairie dogs. Ecology. 1995;76:1794–1806. [Google Scholar]
- Voigt CC, von Helversen O. Storage and display of odor by male Saccopteryx bilineata (Chiroptera; Emballonuridae) Behav Ecol Sociobiol. 1999;47:29–40. [Google Scholar]
- Wedekind C, Chapuisat M, Macas E, Rülicke T. Non-random fertilization in mice correlates with the MHC and something else. Heredity. 1996;77:400–409. doi: 10.1038/hdy.1996.160. [DOI] [PubMed] [Google Scholar]
- Wedekind C, Seebeck T, Bettens F, Paepke AJ. MHC-dependent mate preferences in humans. Proc R Soc Lond B. 1995;260:245–249. doi: 10.1098/rspb.1995.0087. [DOI] [PubMed] [Google Scholar]
- Wegner KM, Kalbe M, Kurtz J, Reusch TBH, Milinski M. Parasite selection for immunogenetic optimality. Science. 2003;301:1343. doi: 10.1126/science.1088293. [DOI] [PubMed] [Google Scholar]
- Wibbelt G, Moore M, Schountz T, Voigt CC. Emerging diseases in Chiroptera: why bats. Biol Lett. 2010;6:438–440. doi: 10.1098/rsbl.2010.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfig B, Traulson A, Milinski M, Boehm T. Does intra-individual major histocompatibility complex diversity keep a golden mean. Phil Trans R Soc B. 2009;364:117–128. doi: 10.1098/rstb.2008.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Lau S, Woo P, Yuen K-Y. Bats as a continuing source of emerging infections in humans. Rev Med Virol. 2007;17:67–91. doi: 10.1002/rmv.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WSW, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identification of positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington Wilmer J, Barrat E. A non-lethal method of tissue sampling for genetic studies of chiropterans. Bat Res News. 1996;37:1–3. [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]
- Yang ZH. PAML: a program for package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;15:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Ziegler A, Dohr G, Uchanska-Ziegler B. Possible roles for products of polymorphic MHC and linked olfactory receptor genes during selection processes in reproduction. Am J Reprod Immunol. 2002;48:34–42. doi: 10.1034/j.1600-0897.2002.01097.x. [DOI] [PubMed] [Google Scholar]
- Ziegler A, Kentenich H, Uchanska-Ziegler B. Female choice and the MHC. Trends Immunol. 2005;26:496–502. doi: 10.1016/j.it.2005.07.003. [DOI] [PubMed] [Google Scholar]