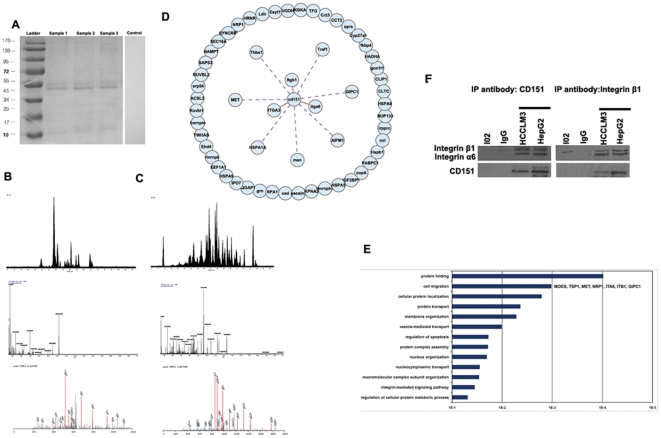
Figure 1. Identification of tetraspanin CD151 network and functional analysis.
CD151-containing complexes were solubilized using the mild detergent, Brij97, and were isolated by immunoprecipitation using specific CD151 mAbs. Associated proteins were eluted using Triton X-100 and separated by SDS-PAGE (A). After in-gel trypsin digestion of proteins, the resulting peptides were analyzed using LC-MS/MS (B and C). The peptides were separated by nano-HPLC. Total ion count was measured and visualized on a chromatogram (upper panel). At a precise time (e.g. 55.33 min for CD151 and 67.18 min for integrin β1, dotted straight line), the mass spectrum obtained is shown (middle panel) in which a parent ion can be selected (e.g. m/z = 643.3489 for CD151 and 1010.0838 for integrin β1, black arrow). Fragmentation of that parent ion led to MS/MS spectrum generation containing b and y ions, and thus sequence information of the parent ion (lower panel). The amino acid sequence can be deduced after searches in the NCBI database using the program SEQUEST. The putative sequence of the peptide is shown with associated Xcorr and ΔCn. This peptide sequence led to the identification of CD151 and integrin β1. These peptides were converted into a gene symbol and rearranged according to GO functional enrichment provided by DAVID website. Then, the “tetraspanin CD151 web” was organized by the cytoscape (Ver: 2.6.2, http://www.cytoscape.org/), searching target genes that interacted with CD151, and removing genes that were not contained in our target genes using a data source (NCBI Entrez EUtilities Web Service Client) (D). Gene ontology analysis of the 57 molecular partners was performed. They were classified as 13 categories of biological processes including protein folding and cell migration (MOES, TSP1, c-MET, NRP1, ITA6, ITB1 and GIPC1) according to categories of the “GO Biological Process” (E). Western blotting identified CD151 formed complexes with integrin α6 and β1 in HCCLM3 cells as well as HepG2 cells, but not in normal hepatocytes L-02 (F).