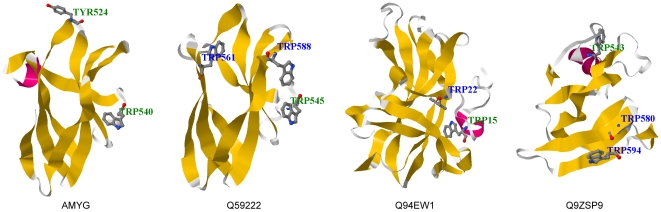
Figure 1. CBMs containing known ligand-binding aromatic residues and their in silico structures.
The four selected CBMs with experimentally determined ligand-binding aromatic residues are Aspergillus oryzae glucoamylase (AMYG, CBM20, [22]), Bacillus sp. TS-23 α-amylase (Q59222, CBM20, [23]), Nicotiana tabacum Nictaba (Q94EW1, CBM22, [24]), and Solanum lycopersicum endo-β-1,4-D-glucanase (Q9ZSP9, CBM49, [25]). Known ligand-binding aromatic residues are highlighted in ball and stick model. HARs and non-HARs are texted in green and blue, respectively. Five out of ten known ligand-binding residues are predicted as HARs. The 3D structures were rendered by Jmol (http://www.jmol.org/).