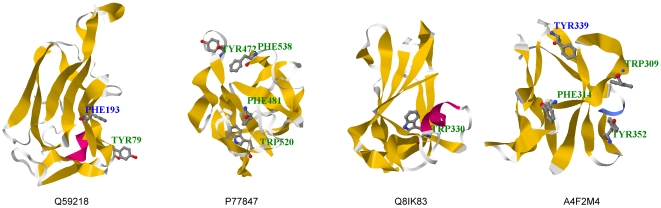
Figure 2. CBMs containing aromatic residues conserved to known ligand-binding residues and their in silico structures.
The four selected CBMs with aromatic residues conserved to known ligand-binding residues are Bacteroides ovatus arabinosidase (Q59218, CBM4), Caldocellum saccharolyticum β-1,4-annanase (P77847, CBM6), Plasmodium falciparum LCCL domain-containing protein (Q8IK83, CBM32), and Phaseolus vulgaris starch synthase III (A4F2M4, CBM53). Known ligand-binding aromatic residues are highlighted in ball and stick model. HARs and non-HARs are texted in green and blue, respectively. Nine out of eleven aromatic residues conserved to known ligand-binding residues are predicted as HARs. The 3D structures were rendered by Jmol (http://www.jmol.org/).