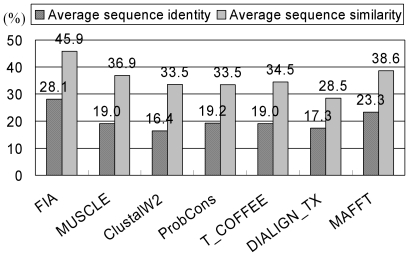
Figure 3. Average sequence identities and similarities for CBMs from seven sequence alignment methods.
The sequence identities (similarities) are averaged from 75,981 (817 * 93) target-template pairwise sequence alignments. The used alignment programs are FIA [21], MUSCLE [32], ClustalW2 [33], ProbCons [36], T-COFFEE [35], DIALIGN-TX [34], and MAFFT [37].