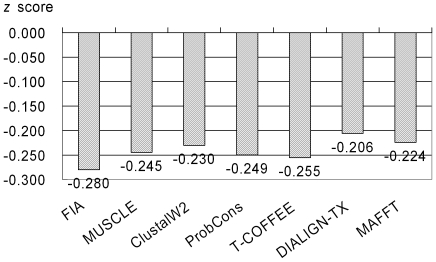
Figure 4. Average z scores for in silico structures built based on seven sequence alignment methods.
The z scores are averaged from the lowest z scores of top five in silico structures for each target CBM. The used alignment programs are FIA [21], MUSCLE [32], ClustalW2 [33], ProbCons [36], T-COFFEE [35], DIALIGN-TX [34], and MAFFT [37]. The surface potential z scores are evaluated by PROSA2003 [38].