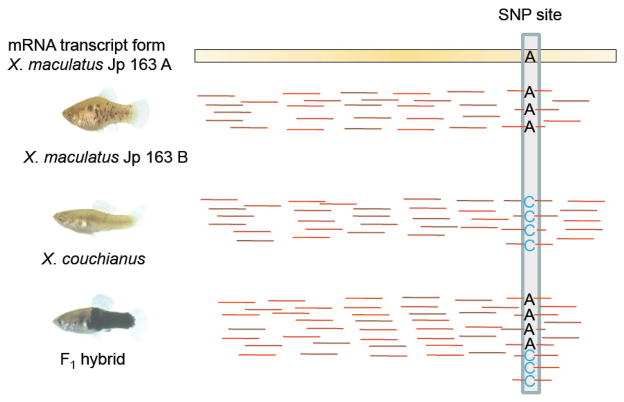
Figure 2.
A diagrammatic example of identification of SNPs and measurement of ASGE in F1 interspecies hybrids. Red bars represent RNA-Seq reads mapped to the reference transcriptome. Most reads from X. maculatus Jp 163 B match perfectly to the Jp 163 A reference transcriptome. RNA-seq reads from X. couchianus were also mapped to X. maculatus Jp 163 A reference transcriptome and SNPs sites were identified by comparing consensus bases of RNA-seq reads (C in this case) to the corresponding base in the reference transcriptome (A in this case). In the hybrid, reads mapped to SNPs sites are classified by the bases they carry and counted separately as the measurement of ASGE. In this SNP, 4 X. maculatus allele reads and 3 X. couchianus allele reads were counted in the hybrid.