Abstract
This mini-review focuses on a knockdown technology called deoxyribozymes, which has rarely been utilized in the field of neurobiology/neuroscience. Deoxyribozymes are catalytic DNA molecules, which are also entitled DNA enzyme or DNAzyme. This mini-review presents a description of their development, structure, function, and therapeutic application. In addition, information on siRNA, ribozymes, and antisense are given. Further information on two deoxyribozymes against c-Jun and xylosyltransferase (XT) mRNA are summarized of which the first is important to influence many neurological disorders and the last potentially treats spinal cord injuries (SCIs). In particular, insults to the central nervous system (CNS) such as SCI generate an inhibitory environment (lesion scar) at the injury site that prevents the endogenous and therapy-induced axonal regeneration and thereby limits repair strategies. Presently, there are no treatments available. Hence, deoxyribozymes provide an opportunity for new therapeutics that alter the inhibitory nature of the lesion scar and thus promote axonal growth in the injured spinal cord. When used cautiously and within the limits of its ability the deoxyribozyme technology holds promise to become a major contributing factor in repair strategies of the CNS.
Keywords: central nervous system trauma, brain insult, drug development, DNA enzymes, catalytic DNA, c-Jun, xylosyltransferase, proteoglycans
Introduction
Scientists have worked for over 100 years to understand the mechanisms that are involved during axonal growth and regeneration processes. In spite of considerable success no medication is available. Hence, the development of therapeutics to treat central nervous system (CNS) trauma such as spinal cord injury (SCI) is a long awaited intervention in pursuit of finding a cure for paralysis. Presently, approximately 10,000 people sustain a SCI every year in Europe; whereas approximately 40 million patients worldwide are paralyzed due to damage to their spinal cord. About 70% of all these victims sustained a contusive injury. Most of them are young men with an average age of 33.4. As a consequence about 2.5 million people must presently live wheelchair-bound or they are even worse off (Adams and Cavanagh, 2004). An early treatment to prevent long-term complications after SCI with the goal to accelerate recovery is a necessary step toward medicating CNS trauma. Hence, this review will introduce the deoxyribozyme technology as a potential therapeutic that promotes regeneration and axonal growth after insults to the brain or spinal cord. Its development, structure, function, design, and therapeutic applications will be reviewed to provide an alternative to common knockdown agents.
Deoxyribozyme
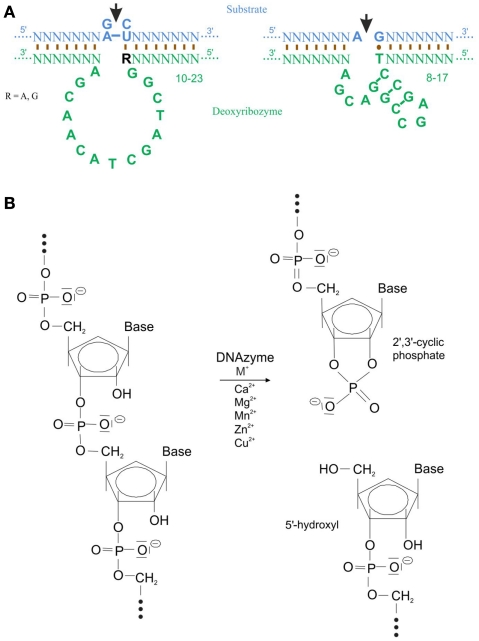
Single-stranded, catalytic DNA molecules, which are also being called DNA enzymes or DNAzymes, have been reported not to exist in nature. Instead they have been developed by Santoro and Joyce (1997) who used a technique called systematic evolution of ligands by exponential enrichment (SELEX). Here, a selection process was used, in which a RNA substrate was provided to a library of approximately 1014 potential single-stranded DNA enzymes. Via enrichment of the functional catalytic DNA molecules two deoxyribozymes named 10-23 and 8-17 with a length of approximately 35 nucleotides were isolated (for review, Silverman, 2005). The catalytic loop structure of the 10-23 deoxyribozyme contains 13, the one of the 8-17 15 nucleotides, of which each is surrounded by sequence-specific binding arms (Figure 1A). To be able to digest any target mRNA and hence prevent the translation to a protein these two deoxyribozymes require divalent metal ions such as Ca2+, Mg2+, Mn2+, Zn2+, or Cu2+. These metals function as a general base or as a Lewis acid either to stabilize the transition state of the reaction and/or to assist in folding of the enzyme into an active conformation (Santoro and Joyce, 1998). An exception is the deoxyribozyme motif, Na8, which increases RNA cleavage by 108-fold in the absence of divalent metals (Geyer and Sen, 1997). Another deoxyribozyme requires l-histidine or a closely related analog to catalyze RNA phosphoester cleavage (Roth and Breaker, 1998). Target specificity of the deoxyribozyme is controlled by the sequence of the binding arms. The binding arms of both deoxyribozymes form Watson–Crick base pair interactions of 8–12 nucleotides with their target mRNA. A phosphodiester linkage in this target mRNA is cleavage by deoxyribozymes between an unpaired purine (adenine [A] or guanine [G]) and a paired pyrimidine (cytosine [C] or uracil [U]) to an end product of 2′,3′-cyclic phosphate and 5′-hydroxyl RNA termini (Figure 1B). In case of the 10-23 deoxyribozyme the required dinucleotides in the target mRNA are G-C, G-U, A-C, or A-U whereas the 8-17 deoxyribozyme requires solely A-G as recognition sequence. The order of best to worst substrate cleavage is as follow: rAU = rGU ≥ rGrC ≫ rArC, whereas the relatively poor activity of a deoxyribozyme against rArC- and rGrC-containing substrate can substantially be increased by chemical modifications to the binding domain that subtly weaken the interaction with the substrate. When dG is substituted with deoxyinosine (dI) such that the three hydrogen bonds between rC–dG are converted to two hydrogen bonds rC–dI, the biological activity of that deoxyribozymes that target rRrC junctions (R = rA, rG) in the substrate is improved (Cairns et al., 2003). To prevent deoxyribozyme digestion through serum nucleases, the following modifications are the most frequently used: (1) phosphorothioated (PS) bases at the 5′ and/or 3′ end; (2) 3′-3′-linked inverted thymidine or any other inverted nucleotide; (3) locked nucleic acid (LNA); or (4) phosphoramidated bases (for more detail on the design of deoxyribozymes please see Achenbach et al., 2004).
Figure 1.
Structure and digestion mechanism of deoxyribozymes. (A) Structure of the 10-23 and 8-17 deoxyribozyme. The catalytic loop digest the target mRNA (marked by the arrow) between an unpaired purine and a paired pyrimidine nucleotide. Binding to the mRNA occurs mostly via Watson–Crick base pairing. Note a G-T Hoogsteen base pair in the 8-17 deoxyribozyme (modified from Santoro and Joyce, 1997). (B) Transesterification of a target mRNA via a deoxyribozyme and divalent cations generates a 2′3′-cyclic phosphate and a 5′-hydroxyl termini as end product (modified from Achenbach et al., 2004).
Deoxyribozymes show high flexibility, specificity, and low production costs, which makes them a valuable asset for treatment of, e.g., SCI. In addition they have the advantage to be administered and internalized into cells without using transfection agents, electroporation, or infection albeit utilizing a concentration that does not induce side effects. Hence, any mRNA that participates in the inhibitory environment after insult to the CNS can serve as a substrate for digestion. The utilization of deoxyribozymes alone or in combination with other approaches has the potential to generate a growth-permissive environment at the lesion site. This holds promise to enhance axonal regeneration, and thus, increases the chances of functional recovery.
Other Knock Down Technologies
Small interfering RNA
Presently, siRNA is the most frequently used knockdown technique in science with developments of approximately 21 potential therapeutics to a variety of diseases (Burnett et al., 2011). The mechanism of siRNA, which are double-stranded RNA molecules of usually 21 nucleotides that contain a 3′ overhang of 2 nucleotide on either end, is well known and documented by a plethora of publications (see reviews: Fire, 1999; Tuschl, 2001; Mello and Conte, 2004). In addition to its role to interfere with the expression of specific genes, it also acts in antiviral mechanisms or shaping the chromatin structure in a genome. siRNA was discovered by Andrew Fire and Craig Mello, who received the Nobel Prize 2006. In light of such excitement and success it is surprising that Roche closed, at the end of last year, all R&D facilities that were working on identifying siRNAs sequences to treat various diseases and disorders. In addition Novartis terminated a contract with Alnylam, a company that provides a platform for the identification of siRNA target sequences. However, Novartis continues to work on the 31 siRNAs, which they have internally but they are not interested in indentifying new sequences. Furthermore, Pfizer and Abbott also demonstrate no further interest in this technology. Instead, Pfizer (New York) in conjunction with Santaris (Horsholm, Denmark) formed an alliance to develop an antisense strategy utilizing PS–LNAs (Pollack, 2011; Schmidt, 2011).
Ribozymes
Ribozymes (RZs), mostly used as hammerhead or hairpin RZs in therapeutic applications, are naturally occurring, single-stranded RNA molecules, which contain a catalytic loop structure and cleave their target RNA in a sequence-specific manner. They have been discovered in the 1980s for which Sidney Altman and Thomas Cech received the Nobel Prize 1989. RZs were the first demonstration that RNA is able to have catalytic/enzymatic properties, which lead to the belief that DNA molecules in form of deoxyribozymes are also able to digest mRNA. Based on the SciClips database 18 potential drug targets are in focus of RZ research. The recognition sequence of the hammerhead RZs contains a XUN structure, where X is any base and N is A, C, or U (Haseloff and Gerlach, 1988), whereas hairpin RZs recognize RCN/GUCB with B = C, G, or U, N = any base and R = G or A as a target sequence (Joseph et al., 1993).
Antisense
The antisense technology, which was developed in the 1970s (Zamecnik and Stephenson, 1978), is the trailblazer for gene knockdown agents used in science and in particular in the nervous system. Here, a single-stranded DNA molecule, an antisense oligonucleotide (ODN), binds stoichiometrically to the target mRNA and prevents its transcription, splicing, or translation to a protein (Eckstein, 2007). If the cell contains RNAse H, the RNA/DNA hybrid that forms after binding of the antisense ODN to the target pre-mRNA or mRNA is going to be digested. The first antisense drug, Vitravene, which treated cytomegalovirus-based retinitis, was approved 1998 and discontinued in 2004 as the drug’s market shrank (Jones, 2011). Approximately 10 other antisense drugs are being researched. Furthermore, the following proteins c-erbA/rev-erbA (Munroe and Lanzar, 1991), N-myc (Armstrong and Krystal, 1992), gonadotropin releasing hormone (GnRH, Wilson et al., 1995), Wilms’ tumor suppressor gene (WT1, Malik et al., 1995), insulin-like growth factor (IGF-II, Baccarini et al., 1993; Rivkin et al., 1993), proliferating cell nuclear antigen (PCNA, Swalla and Jeffery, 1996), hoxa-11 (Hsieh-Li et al., 1995), and bFGF (FGF-2, Borja et al., 1993) have been observed to be regulated by their natural antisense ODNs. Modifications such as PS oligonucleotides (PTOs), peptide nucleic acids (PNAs), and LNAs as well as developments of morpholinos have their origin during antisense development.
Therapeutic Applications of Deoxyribozymes
Only the 10-23 deoxyribozyme, in particular, with kcat and KM values comparable to those of naturally occurring ribozymes has been applied in vivo to inactivate target mRNA molecules (Santoro and Joyce, 1997). As Table 1 demonstrates, 10 in vivo applications of deoxyribozymes to various target mRNAs exist, however, their utilization in the neurobiology field is quite limited. Therefore, to focus on rather CNS related applications the mini-review will focus on c-Jun and xylosyltransferase-1 (XT-1) deoxyribozyme applications in vivo.
Table 1.
In vivo application of deoxyribozymes (modified from Isaka, 2007).
| Gene target | Disease (tissue) | Application | Reference |
|---|---|---|---|
| GENERAL | |||
| Erg-1 | Balloon injury (carotid artery) | Adventitial (from outside of a vessel) | Santiago et al. (1999) |
| Ligation (carotid artery) | Adventitial | Lowe et al. (2002) | |
| Restenosis (coronary artery) | Endoluminal (from inside a vessel) | Lowe et al. (2001) | |
| Tumor growth (breast carcinoma) | Transfection, subcutane, intratumoral | Fahmy et al. (2003), Mitchell et al. (2004) | |
| Ureteral obstruction (kidney) | Interstitial | Nakamura et al. (2002) | |
| Myocardial infarction | Intramyocardial | Bhindi et al. (2006) | |
| TGF-β | Glomerulonephritis | Renal arterial | Isaka et al. (2004) |
| c-Jun | Neovascularization (cornea) | Injection, intravitreal | Zhang et al. (2004), Khachigian et al. (2002) |
| Tumor growth (melanoma) | Subcutane | Zhang et al. (2004) | |
| Tumor growth (squamous cell carcinoma) | Transfection | Zhang et al. (2006) | |
| Inflammatory diseases | Intravitreal, intradermal, topical | Fahmy et al. (2006) | |
| VEGFR-2 | Tumor growth (breast carcinoma) | Intratumoral | Zhang et al. (2002) |
| TNF-α | Myocardial infarction | Intraperitoneal | Iversen et al. (2001) |
| VDUP1 | Myocardial ischemia | Intracardiac | Xiang et al. (2005) |
| PAI-1 | Myocardial infarction | Intracardiac | Xiang et al. (2004) |
| GATA-3 | Asthma (TH2 cells, mast cells, eosinophils, and epithelial cells) | Intranasal/topical | Sel et al. (2008) |
| ftsZ | Sepsis (bacterial cell division) | Systemic | Tan et al. (2004) |
| NEUROSCIENCE | |||
| Xylosyltransferase-1 (XT-1) | Spinal cord injury | Intrathecal | Grimpe and Silver (2004), Hurtado et al. (2008) |
c-Jun is a basic region leucine-zipper transcription factor that forms homo- and heterodimers, latter in combination with c-fos, ATF, CREB, steroid hormone receptor, or JDP members. It binds to the AP-1, CRE, or NF-AT consensus sequence in promoter regions of immediate early genes (see review: Antoniou et al., 2011). The transcription factor is not only essential to neuronal differentiation but is also a powerful mediator of neuronal apoptotic signal cascades involving c-Jun N-terminal kinases (JNKs, Kyriakis et al., 1994). In addition, JNK activation is strongly involved in inflammatory responses, neurodegeneration, and in the development of hyperalgesia and allodynia (see mini-review: Gao and Ji, 2008). Hence, the JNK pathway is an important therapeutic target to influence diverse neurological disorders (see review: Antoniou et al., 2011). Investigations by Dass and Choong (2010) showed that the intravenous (iv) administration of a deoxyribozyme to c-Jun mRNA (Dz13) in a bolus dose or in an in utero assay has no toxic effect to blood and solid tissues in adult or fetal mice, only a slight hepatotoxicity was noted with histology.
After insult to the CNS, e.g., SCI an axonal growth inhibitory environment forms at the injury site. Proteoglycans (PGs) such as chondroitin sulfate (CS−) PGs constitute a major group of molecules that participates in this axon growth-inhibition. They most likely exert their inhibitory effects through glycosaminoglycan (GAG)-side chains. GAG-chain synthesis is initiated through an enzyme called XT-1, which attaches a xylose to the PG core protein. Application of a deoxyribozyme to XT-1 not only prevents glycosylation of PGs but also avoids the assembly of the PG core protein into the extracellular matrix. As a consequence, microtransplanted and endogenous severed sensory axons are able to grow beyond the lesion site because the inhibitory substrate is reduced (Grimpe and Silver, 2004; Grimpe et al., 2005; Hurtado et al., 2008). Hence, the deoxyribozyme to XT-1 mRNA holds promise to be a contributing factor in treating CNS injuries.
Whether deoxyribozymes induce immunostimulatory activity requires elucidation. Characteristics of single-stranded DNA molecules to activate such innate immunity are the presence of a partially (7–10 nucleotides) or completely PS backbone, a poly G tail at the 3′ end, 5′ end, or both, an internal palindrome sequence and an unmethylated CpG motif. CpG motifs induce splenic B cell proliferation, dendritic cell maturation, antibody production, activation of T lymphocytes and natural killer cells as well as cytokine production such as interleukin (IL)-6, IL-12, and interferon Γ. The CpG motif is considered a pathogen-associated molecular pattern, which is recognized by Toll-Like Receptor 9 (TLR9). TLR9 is constitutively expressed only in B cells and plasmacytoid dendritic cells in humans and other higher primates (Krug et al., 2001). Such CpG motifs are likely to occur in deoxyribozymes. However, no information is available on the immunological status of deoxyribozymes. Explicit information on the half-life of deoxyribozymes in the cell or after systemic administration is also not available. However, extensive investigations on antisense PTOs revealed that their plasma half-life after iv administration lies between 30–85 min (Stevenson et al., 1999, see review: Wang et al., 2001) whereas within the cell contains their lifespan approximately 2–3 days.
The steady success of the deoxyribozyme technology to generate potential therapeutics funneled into the foundation of two start up companies, the US-based CytoGenix Inc. (Tan et al., 2004) and the Germany-based Sterna Biologicals (Sel et al., 2008), which tests DNA enzymes in preclinical trials of cancer, sepsis, and asthma research.
Conclusion
All above mentioned knockdown technologies are incapable to pass the blood–brain or spinal cord-barrier based on their charged DNA or RNA backbone. Modifications such as PTOs, PNA, and LNA are utilized to make up for this deficit. In addition, they prevent exonuclease degradation and increase stability. To direct any of the molecules to specific regions of the body, e.g., a tumor, is an equal challenge to all aforementioned knockdown agents, whereas uptake into cells without any aid is in favor of single-stranded molecules. All together one can say each of the knockdown techniques has its pros and cons depending on scientific demands or personal preference. Hence, each agent needs to be evaluated from case to case. There might not even be one technology to favor but at the same time, none of these techniques should be overlooked either.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research on deoxyribozymes in the Grimpe laboratory has been supported by the German Research Foundation (DFG), Forschungskommission der HHU, Wilson Medical Research Foundation, and Glaucoma Foundation.
Abbreviations
AP, activator protein; ATF, activating transcription factor; CNS, central nervous system; CRE, cAMP responsive element; CREB, cAMP response element binding protein; CS, chondroitin sulfate; DNA, deoxyribonucleic acid; Dz, deoxyribozyme; FGF, fibroblast growth factor; GAG, glycosaminoglycan; GnRH, gonadotropin releasing hormone; IGF, insulin-like growth factor; IL, interleukin; iv, intravenous; JDP, Jun dimerization protein; JNK, c-Jun N-terminal kinase; LNA, locked nucleic acid; NF-AT, nuclear factor of activated T-cells; ODN, oligonucleotide; PCNA, proliferating cell nuclear antigen; PG, proteoglycan; PNA, peptide nucleic acid; PS, phosphorothioated; PTO, PS oligonucleotide; R&D, research and development; RNA, ribonucleic acid; RZ, ribozyme; SCI, spinal cord injury; SELEX, systematic evolution of ligands by exponential enrichment; siRNA, small interfering RNA; TLR, toll-like receptor; US, United States; WT, Wilms’ tumor; XT, xylosyltransferase.
References
- Achenbach J. C., Chiuman W., Cruz R. P. G., Li Y. (2004). DNAzymes: from creation in vitro to application in vivo. Curr. Pharm. Biotechnol. 5, 321–336 10.2174/1389201043376751 [DOI] [PubMed] [Google Scholar]
- Adams M., Cavanagh J. F. (2004). International Campaign for Cures of Spinal Cord Injury Paralysis (ICCP): another step forward for spinal cord injury research. Spinal Cord 42, 273–280 10.1038/sj.sc.3101597 [DOI] [PubMed] [Google Scholar]
- Antoniou X., Falconi M., DiMarino D., Borsello T. (2011). JNK3 as a therapeutic target for neurodegenerative diseases. J. Alzheimers Dis. 24, 633–642 [DOI] [PubMed] [Google Scholar]
- Armstrong B. C., Krystal G. W. (1992). Isolation and characterization of complementary DNA for N-cym, a gene encoded by the DNA strand opposite to N-myc. Cell Growth Differ. 3, 385–390 [PubMed] [Google Scholar]
- Baccarini P., Fiorentino M., D’Errico A., Mancini A. M., Grigioni W. F. (1993). Detection of anti-sense transcripts of the insulin-like growth factor-2 gene in Wilms’ tumor. Am. J. Pathol. 143, 1535–1542 [PMC free article] [PubMed] [Google Scholar]
- Bhindi R., Khachigian L. M., Lowe H. C. (2006). DNAzymes targeting the transcription factor Egr-1 reduce myocardial infarct size following ischemia-reperfusion in rats. J. Thromb. Haemost. 4, 1479–1483 10.1111/j.1538-7836.2006.02022.x [DOI] [PubMed] [Google Scholar]
- Borja A. Z. M., Meijers C., Zeller L. (1993). Expression of alternatively spliced bFGF first coding exons and antisense mRNAs during chicken embryogenesis. Dev. Biol. 157, 110–118 10.1006/dbio.1993.1116 [DOI] [PubMed] [Google Scholar]
- Burnett J. C., Rossi J. J., Tiemann K. (2011). Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol. J. 6, 1130–1146 10.1002/biot.201100054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns M. J., King A., Sun L. Q. (2003). Optimization of the 10-23 DNAzyme-substrate pairing interactions enhanced RNA cleavage activity at purine-cytosine target sites. Nucleic Acids Res. 31, 2883–2889 10.1093/nar/gkg378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dass C. R., Choong P. F. M. (2010). Sequence-related off-target effect of Dz13 against human tumor cells and safety in adult and fetal mice following systemic administration. Oligonucleotides 20, 51–60 10.1089/oli.2009.0226 [DOI] [PubMed] [Google Scholar]
- Eckstein F. (2007). The versatility of oligonucleotides as potential therapeutics. Expert Opin. Biol. Ther. 7, 1021–1034 10.1517/14712598.7.7.1021 [DOI] [PubMed] [Google Scholar]
- Fahmy R. G., Dass C. R., Sun L. Q., Chesterman C. N., Khachigian L. M. (2003). Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat. Med. 9, 1026–1032 10.1038/nm905 [DOI] [PubMed] [Google Scholar]
- Fahmy R. G., Waldman A., Zhang G., Mitchell A., Tedla N., Cai H., Geczy C. R., Chesterman C. N., Perry M., Khachigian L. M. (2006). Suppression of vascular permeability and inflammation by targeting of the transcription factor c-Jun. Nat. Biotechnol. 24, 856–863 10.1038/nbt1225 [DOI] [PubMed] [Google Scholar]
- Fire A. (1999). RNA-triggered gene silencing. Trends Genet. 15, 358–363 10.1016/S0168-9525(98)01650-3 [DOI] [PubMed] [Google Scholar]
- Gao Y. J., Ji R. R. (2008). Activation of JNK pathway in persistent pain. Neurosci. Lett. 6, 180–183 10.1016/j.neulet.2008.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer C. R., Sen D. (1997). Evidence for the metalcofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol. 4, 579–593 10.1016/S1074-5521(97)90244-1 [DOI] [PubMed] [Google Scholar]
- Grimpe B., Pressman Y., Bunge M. B., Silver J. (2005). The role of proteolgycans in Schwann cell/astrocyte interactions and in regeneration failure at PNS/CNS interfaces. Mol. Cell. Neurosci. 28, 18–29 10.1016/j.mcn.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Grimpe B., Silver J. (2004). A novel DNA enzyme reduces glycosaminoglycan chains in the gliar scar and allows microtransplanted dorsal root ganglia axons to regenerate beyond lesions in the spinal cord. J. Neurosci. 24, 1393–1397 10.1523/JNEUROSCI.4986-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. (1988). Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature 334, 585–591 10.1038/334585a0 [DOI] [PubMed] [Google Scholar]
- Hsieh-Li H. M., Witte D. P., Weinstein M., Branford W., Li H., Small K., Potter S. S. (1995). Hoxa 11 structure, extensive antisense transcription, and function in male and female fertility. Development 121, 1373–1385 [DOI] [PubMed] [Google Scholar]
- Hurtado A., Podini H., Oudega M., Grimpe B. (2008). Deoxyribozyme-mediated knock down of xylosyltransferase-1 mRNA promotes sensory axon regeneration in the adult rat spinal cord. Brain 131, 2596–2605 10.1093/brain/awn206 [DOI] [PubMed] [Google Scholar]
- Isaka Y. (2007). DNAzymes as potential therapeutic molecules. Curr. Opin. Mol. Ther. 9, 132–139 [PubMed] [Google Scholar]
- Isaka Y., Nakamura H., Mizui M., Takabatake Y., Horio M., Kawachi H., Shimizu F., Imai E., Hori M. (2004). DNAzyme for TGF-β suppressed extracelluar matrix accumulation in experimental glomerulonephritis. Kidney Int. 66, 586–590 10.1111/j.1523-1755.2004.00777.x [DOI] [PubMed] [Google Scholar]
- Iversen P. O., Nicolaysen G., Sioud M. (2001). DNA enzyme targeting TNF-α mRNA improves hemodynamic performance in rats with postinfarction heart failure. Am. J. Physiol. Heart Circ. Physiol. 281, H2211–H2217 [DOI] [PubMed] [Google Scholar]
- Jones D. (2011). The long march of antisense. Nat. Rev. Drug Discov. 10, 401–402 10.1038/nrd3353 [DOI] [PubMed] [Google Scholar]
- Joseph S., Berzal-Herranz A., Chowrira B. M., Butcher S. E., Burke J. M. (1993). Substrate selection rules for the hairpin ribozyme determined by in vitro selection, mutation, and analysis of mismatched substrates. Genes Dev. 7, 130–138 10.1101/gad.7.1.130 [DOI] [PubMed] [Google Scholar]
- Khachigian L. M., Fahmy R. G., Zhang G., Bobryshev Y. V., Kaniaros A. (2002). c-Jun regulates vascular smooth muscle cell growth an neointima formation after arterial injury: inhibition by a novel DNAzyme targeting c-Jun. J. Biol. Chem. 277, 22985–22991 10.1074/jbc.M200977200 [DOI] [PubMed] [Google Scholar]
- Krug A., Rothenfusser S., Hornung V., Jahrsdörfer B., Blackwell S., Ballas Z. K., Endres S., Krieg A. M., Hartmann G. (2001). Identification of CpG oligonucleotide sequences with high induction of IFN-alpha/beta in plasmacytoid dendritic cells. Eur. J. Immunol. 31, 2154–2163 [DOI] [PubMed] [Google Scholar]
- Kyriakis J. M., Baneriee P., Nikolakaki E., Dai T., Rubie E. A., Ahma D. M. F., Avruch J., Woodgett J. R. (1994). Teh stress-activated protein kinase subfamily of c-Jun kinase. Nature 369, 156–160 10.1038/369156a0 [DOI] [PubMed] [Google Scholar]
- Lowe H. C., Chesterman C. N., Khachigian L. M. (2002). Catalytic antisense DNA molecules targeting Egr-1 inhibit neointima formation following permanent ligation of rat common carotid arteries. Thromb. Haemost. 87, 134–140 [PubMed] [Google Scholar]
- Lowe H. C., Fahmy R. G., Kavurna M. M., Baker A., Chesterman C. N., Khachigian L. M. (2001). Catalytic oligodeoxynucleotides define a key regulatory role for early growth response factor-1 in the porcine model of coronary in-stent restenosis. Circ. Res. 89, 670–677 10.1161/hh2001.097867 [DOI] [PubMed] [Google Scholar]
- Malik K. T. A., Wallace J. I., Ivins S. M., Brown K. W. (1995). Identification of an antisense WT1 promoter in intron 1: implications for WT1 gene regulation. Oncogene 11, 1589–1595 [PubMed] [Google Scholar]
- Mello C. C., Conte D., Jr. (2004). Revealing the world of RNA interference. Nature 431, 338–342 10.1038/nature02872 [DOI] [PubMed] [Google Scholar]
- Mitchell A., Dass C. R., Sun L. Q., Khachigian L. M. (2004). Inhibition of human breast carcinoma proliferation, migration, chemoinvasion and solid tumor growth by DNAzymes targeting the zinc finger transcription factor EGR-1. Nucleic Acids Res. 32, 3065–3069 10.1093/nar/gkh626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe S. H., Lanzar M. A. (1991). Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J. Biol. Chem. 266, 22083–22086 [PubMed] [Google Scholar]
- Nakamura H., Isaka Y., Tsujie M., Rupprecht H. D., Akagi Y., Ueda N., Imai E., Hori M. (2002). Introduction of DNA enzyme for Egr-1 into tubulointerstitial fibroblasts by electroporation reduced interstitial α-smooth muscle actin expression and fibrosis in unilateral ureteral obstruction (UUO) rats. Gene Ther. 9, 495–502 10.1038/sj.gt.3301724 [DOI] [PubMed] [Google Scholar]
- Pollack A. (2011). Drugmakers’ fever for the power of RNA Interference has cooled. New York Times, 7th February. Available at: http://www.nytimes.com/2011/02/08/science/08rna.html?_r = 2
- Rivkin M., Rosen K. M., Villa-Komaroff L. (1993). Identification of an antisense transcript from the IGF-II locus in mouse. Mol. Reprod. Dev. 35, 394–397 10.1002/mrd.1080350413 [DOI] [PubMed] [Google Scholar]
- Roth A., Breaker R. D. (1998). An amino acid as a cofactor for a catalytic polynucleotide. Proc. Natl. Acad. Sci. U.S.A. 95, 6027–6031 10.1073/pnas.95.11.6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago F. S., Lowe H. C., Kavurma M. M., Chesterman C. N., Baker A., Atkins D. G., Khachigian L. M. (1999). New DNA enzyme targeting Erg-1 mRNA inhibits vascular smooth muscle proliferation and regrowth after injury. Nat. Med. 5, 1264–1269 10.1038/71020 [DOI] [PubMed] [Google Scholar]
- Santoro S. W., Joyce G. F. (1997). A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U.S.A. 94, 4262–4266 10.1073/pnas.94.9.4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro S. W., Joyce G. F. (1998). Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37, 13330–13342 10.1021/bi9812221 [DOI] [PubMed] [Google Scholar]
- Schmidt C. (2011). RNAi momentum fizzles as pharma shifts priorities. Nat. Biotechnol. 29, 93–94 10.1038/nbt0611-469 [DOI] [PubMed] [Google Scholar]
- Sel S., Wegmann M., Dicke T., Sel S., Henke W., Yildirin A. O., Renz H., Garn H. (2008). Effective prevention and therapy of experimental allergic asthma using a GATA-3 specific DNAzyme. J. Allergy Clin. Immunol. 121, 910–916 10.1016/j.jaci.2007.12.1175 [DOI] [PubMed] [Google Scholar]
- Silverman S. K. (2005). Survey and summary: in vitro selection, characterization, and application of deoxyribozymes that cleave RNA. Nucleic Acids Res. 33, 6151–6163 10.1093/nar/gki930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J. P., Yao K. S., Gallagher M., Friedland D., Mitchell E. P., Cassella A., Monia B., Kwoh T. J., Yu R., Holmlund J., Dorr F. A., O’Dwyer P. J. (1999). Phase I clinical/pharmacokinetic and pharmacodynamic trial of the c-raf-1 antisense oligonucleotide ISIS 5132 (CGP 69846A). J. Clin. Oncol. 17, 2227–2236 [DOI] [PubMed] [Google Scholar]
- Swalla B. J., Jeffery W. R. (1996). PCNA mRNA has a 3′UTR antisense to yellow crescent RNA and is localized in ascidian eggs and embryos. Dev. Biol. 178, 23–34 10.1006/dbio.1996.0195 [DOI] [PubMed] [Google Scholar]
- Tan X., Knesha R., Margolin W., Chen Y. (2004). DNA enzyme generated by a novel single-stranded DNA expression vector inhibits expression of the essential bacterial cell division gene ftsZ. Biochemistry 43, 1111–1117 10.1021/bi035164h [DOI] [PubMed] [Google Scholar]
- Tuschl T. (2001). RNA interference and small interfering RNA. Chembiochem 2, 239–245 [DOI] [PubMed] [Google Scholar]
- Wang H., Prasad G., Buolamwini J. K., Zhang R. (2001). Antisense anticancer oligonucleotide therapeutics. Curr. Cancer Drug Targets 1, 177–196 10.2174/1568009013334133 [DOI] [PubMed] [Google Scholar]
- Wilson T. M., Yu-Lee L., Kelley M. R. (1995). Coordinate gene expression of luteinizing hormone-releasing hormone (LHRH) and the LHRH-receptor after prolactin stimulation in the rat Nb2 T-cell line: implications for a role in immunomodulation and cell cycle gene expression. Mol. Endocrinol. 9, 44–53 10.1210/me.9.1.44 [DOI] [PubMed] [Google Scholar]
- Xiang G., Schuster M. D., Seki T., Kocher A. A., Eshghi S., Boyle A., Itescu S. (2004). Down-regualtion of plasminogen activator inhibitor 1 expression promotes myocardial neovascularization by bone marrow progenitors. J. Exp. Med. 200, 1657–1666 10.1084/jem.20040221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang G., Seki T., Schuster M. D., Wittowski P., Boyle A. J., See F., Martens T. P., Kocher A., Sondermejier H., Krum H., Itescu S. (2005). Catalytic degradation of vitamin D up-regulated protein 1 mRNA enhances cardiomyocyte survival and prevents left ventricular remodeling after myocardial ischemia. J. Biol. Chem. 280, 39394–39402 10.1074/jbc.M413116200 [DOI] [PubMed] [Google Scholar]
- Zamecnik P., Stephenson M. (1978). Inhibition of rous sarcoma replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. U.S.A. 75, 280–284 10.1073/pnas.75.1.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Dass C. R., Sumithran E., Di Girolamo N., Sun L. Q., Khachigian L. M. (2004). Effect of deoxyribozymes targeting c-Jun on solid tumor growth and angiogenesis in rodents. J. Natl. Cancer Inst. 96, 683–696 10.1093/jnci/djh120 [DOI] [PubMed] [Google Scholar]
- Zhang G., Luo X., Sumithran E., Pua V. S., Barnerson R. S., Halliday G. M., Khachigian L. M. (2006). Squamous cell carcinoma growth in mice and in culture is regulated by c-Jun and its control of matrix metalloproteinase-2 and -9 expression. Oncogene 25, 7260–7266 10.1038/sj.onc.1209744 [DOI] [PubMed] [Google Scholar]
- Zhang L., Gasper W. J., Stass S. A., Ioffe O. B., Davis M. A., Mixon A. J. (2002). Angiogenic inhibition mediated by a DNAzyme that targets vascular endothelial growth factor receptor 2. Cancer Res. 62, 5463–5469 [PubMed] [Google Scholar]