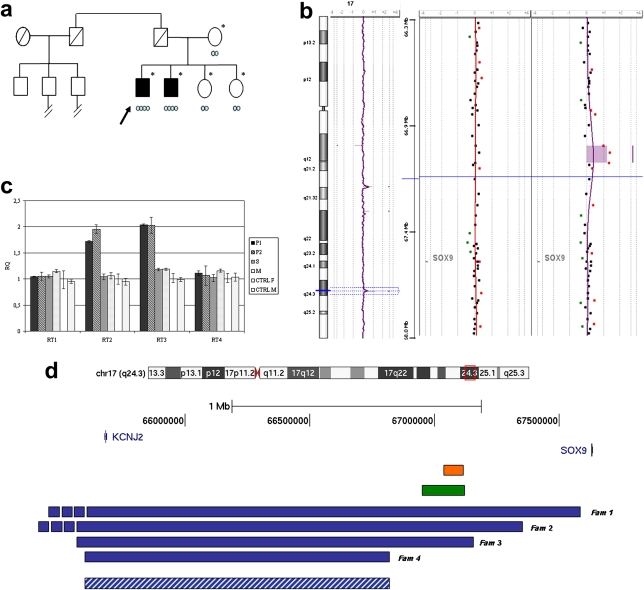
Figure 1.
Family tree of the two XX brothers and characterisation of the 96 kb triplication region. (A) Pedigree of the family. Asterisks indicate persons for whom the karyotype was known, the arrow indicates the proband, and light blue circles indicate the copy number of the 96 kb sex reversal critical region. (B) Array comparative genomic hybridisation (CGH) profile (180K, Agilent Technologies, Santa Clara, California, USA) of the whole chromosome 17 of the proband (left), the 17q24.3 region of the proband (right), and one of his healthy sisters (middle). The triplication is indicated by a shadowed area containing three spots with an average log2 ratio of 1.2. (C) Quantitative PCR (qPCR) with specific primer pairs (supplementary table 3). Affected subjects (P1 and P2), one sister (S) and the mother (M), female (CTRL F) and male (CTRL M) controls were analysed. The relative quantitation (RQ) of copy number is indicated on the y axis. RQ of 1 indicates a normal number of copies; RQ of 2 in P1 and P2 shows the presence of a triplication. Altogether array CGH and qPCR analysis defined the proximal breakpoint of the triplication from 67018227 bp (normal) to 67018939 bp (triplicated) and the distal breakpoint from 67114737 bp (triplicated) to 67119234 bp (normal). Genomic positions are referred to the Human Genome March 2006 (NCBI 36, hg18 assembly). (D) SOX9 and its 1.98 Mb upstream region and a schematic illustration of duplications reported by Kurth et al,4 2009 (blue bars), Cox et al,1 2011 (green bar), and the triplication observed in the present family (orange bar). Minimal critical region for brachydactyly–anonychia (Kurth et al,4 2009) is also shown (blue dashed bar).