Abstract
NhaA, a Na+/H+ antiporter critical for pH and Na+ homeostasis in Escherichia coli, as well as other enterobacteria and possibly Homo sapiens, was modified for fluorescence spectroscopy by constructing a functional Trp-less NhaA mutant. Purified Trp-less NhaA lacks the Trp fluorescence emission characteristic of the wild type, thereby providing a background for studying structure–function relationships in NhaA by site-directed Trp fluorescence. Two single-Trp variants in the Trp-less background (F136W and F339W) were constructed. The mutants grow on selective media, have antiport activities that are similar to Trp-less NhaA, and exhibit Trp fluorescence with three different reversible responses to Li+, Na+, and/or pH. With single Trp/F136W, a pH shift from pH 6.0 to 8.5 induces a red shift and dramatically increases fluorescence in a reversible fashion; no effect is observed when either Na+ or Li+ is added. In marked contrast, with single Trp/F339W, changes in pH do not alter fluorescence, but addition of either Na+ or Li+ drastically quenches fluorescence at alkaline pH. Therefore, a Trp at position 136 specifically monitors a pH-induced conformational change that activates NhaA, whereas a Trp at position 339 senses a ligand-induced conformational change that does not occur until NhaA is activated at alkaline pH.
Keywords: membrane proteins, secondary transporters, transporter functional dynamics
Living cells are critically dependent on processes that regulate intracellular pH, Na+ content, and volume. Na+/H+ antiporters play a primary role in these homeostatic mechanisms (reviewed in refs. 1–3), and such antiporters are found in the cytoplasmic and organelle membranes of most cells (4). Moreover, they have long been human drug targets (5).
NhaA, the principle Na+/H+ antiporter in Escherichia coli, is indispensable for adaptation to high salinity, for challenging Li+ toxicity, and for growth at alkaline pH in the presence of Na+ (1). Several biochemical characteristics of NhaA underpin its physiological roles: very high turnover (6), electrogenicity with a stoichiometry of 2H+/Na+ (1, 7), and a dramatic pH dependence (with maximum activity at alkaline pH), a property it shares with other prokaryotic (1, 6), as well as eukaryotic Na+/H+ antiporters (reviewed in ref. 8).
The recently determined X-ray structure of down-regulated NhaA crystallized at acidic pH (9) provides structural insight into the mechanism of antiport and pH regulation (2). NhaA consists of 12 transmembrane helices (TMs) with the N and C termini on the cytoplasmic side of the membrane. A cytoplasmic funnel opens to the cytoplasm and ends in the middle of the membrane at the putative ion-binding site(s) (9) (Fig. 1A); there is also a periplasmic funnel open to the periplasm and separated from the cytoplasmic funnel by a group of densely packed hydrophobic residues. In addition, the structure reveals a fold in which TMs III, IV, and V are topologically inverted with respect to TMs X, XI, and XII, and in each repeat, one TM (IV and XI, respectively) is interrupted by an extended chain in the middle of the membrane. This noncanonical TM assembly creates a delicately balanced electrostatic environment in the middle of the membrane at the ion-binding site(s), which likely plays a critical role in the cation exchange activity of the antiporter (9). Remarkably, similar structural folds have since been observed in the newly determined structures of other ion-coupled secondary transporters that share little or no sequence homology with NhaA (reviewed in ref. 10). Finally, the structure reveals that NhaA is organized into two functional regions: (i) a cluster of aminoacyl side chains that are involved in pH regulation, which is termed the “pH sensor,” and (ii) a catalytic region containing the ion-binding sites that is 9 Å removed from the pH sensor. Because the structure was determined at pH 4 and NhaA is down-regulated at this pH (9), many questions regarding the active conformation(s) remain.
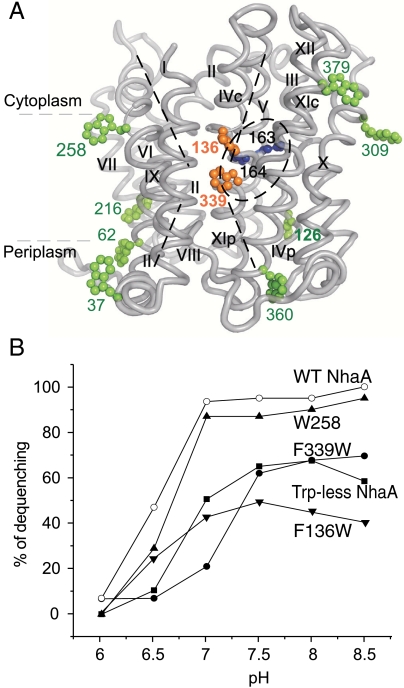
Fig. 1.
NhaA mutations, localization on the crystal structure and Li+/H+ antiporter activity in everted membrane vesicles. (A) The figure (generated using PyMOL) shows the NhaA α-chain and the native Trp (green), the Trp replacements of Phe (orange), and the essential D163 and D164 in the active site (blue), all in ball representation. (B) The Li+/H+ antiporter activity of WT NhaA and the Trp-less variants was determined as in Fig. S1, at the indicated pH values in the presence of 10 mM LiCl. The results are expressed as end level of dequenching (%). All experiments were repeated at least three times with practically identical results.
Changes in fluorescence of native or engineered Trp residues in response to ligand binding or other perturbations have been applied relatively recently to monitor conformational changes in membrane transport proteins (11, 12). Guided by the NhaA crystal structure, we have now applied the approach to examine pH and/or ligand-induced conformational changes in NhaA. The results reveal two conformational changes: one observed at alkaline pH only and independent of ligand, and another that is ligand-induced and observed at alkaline pH only. The former is required for NhaA activation and the latter for antiport activity.
Results
Construction of Trp-Less NhaA.
NhaA has eight native Trp residues, seven of which are located near the phospholipid bilayer and one (Trp258) is in the middle of helix IX (Fig 1A). Although specific and reproducible, the Li+-induced change in Trp fluorescence is very small with purified WT NhaA in dodecyl-β-d-maltopyranoside (DDM) at pH 8.5 (Fig. 2A). This small change is ascribed to a high fluorescence background from most of the native Trp residues, which do not respond to conformational changes, a problem that can be overcome by constructing Trp-less proteins (13). In this regard, replacement of native Trp residues with Tyr rather than Phe was shown to increase expression of Trp-less variants in LacY (14). However, because the Tyr excitation/emission spectra overlap slightly with those of Trp, we chose to engineer a Trp-less NhaA by replacing six Trp (W37, W126, W216, W309, W360, W379) with Phe and two (W62, W258) with Cys (Table S1, plasmid 17). The Cys replacements were shown previously to have WT phenotype (refs. 15 and 16, respectively).
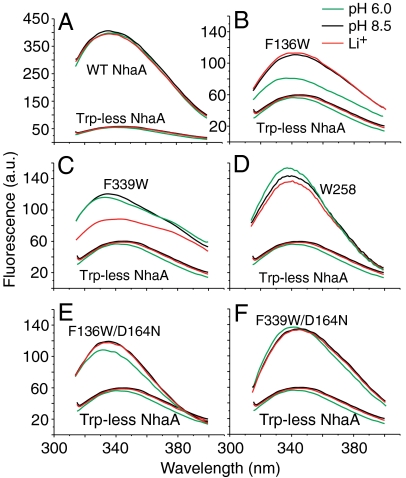
Fig. 2.
Steady-state fluorescence emission spectra of purified WT, Trp-less NhaA, and its variants in DDM micelles and the effect of pH or Li+. Proteins of WT NhaA, Trp-less NhaA, and the indicated single-Trp variants were affinity purified and incubated at protein concentration of 0.3–1 μM in a reaction mixture containing 20 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane at the indicated pH values, 150 mM choline chloride, 5 mM MgCl2, 10% sucrose, and 0.015% DDM at pH 8.5 in the presence (red) or absence (black) of 10 mM LiCl or at pH 6.0 (green) without LiCl. The fluorescence emission spectra were measured with an excitation at 295 nm (A, B, D, F) or at 290 nm (C). All experiments were repeated at least three times using different protein preparations and the results were practically identical.
To characterize the NhaA variants with respect to expression, growth phenotype, and antiporter activity, the mutated plasmids were transformed into EP432, an E. coli strain that lacks the two Na+/H+ antiporters (NhaA and NhaB). This strain neither grows on selective media (0.6 M NaCl at pH 7.0 or 8.2, or 0.1 M LiCl at pH 7.0), nor exhibits any Na+/H+ antiport activity in everted membrane vesicles, unless the parent cells were transformed with a plasmid encoding an active NhaA (reviewed in ref. 1). EP432 transformed with Trp-less NhaA or each of the mutated plasmids grows on both Na+- and Li+-selective agar plates at pH 7, and several including Trp-less NhaA, even grow on Na+-selective plates at pH 8.2 (full stress conditions; ref. 17) similar to the WT control plasmid (S Table 1). Hence, none of the native Trp residues is essential for growth under the respective selective conditions.
Trp-Less NhaA Variants and Growth Phenotype.
Guided by the NhaA structure and in order to minimize structural disturbances, two native Phe residues were replaced with Trp in Trp-less NhaA (F136W or F339W) at sites strategically located with respect to NhaA function (Fig. 1A). Phe136 is located on TM IVc, which lines the cytoplasmic funnel and is in close proximity to helix I, which contributes to the pH sensor (9); Phe339 is located on TM XI in the region of the active site (Fig. 1A). The mutations were also constructed on the WT background (pAXH3/F136W and pAXH3/F339W, Table S1), and native W258 in a Trp-less NhaA background was also constructed (Table S1). W258 is located on TM IX in proximity to the putative pH sensor (2) and faces the NhaA dimer interface (16). Whereas E. coli EP432 expressing either single-Trp/F136W NhaA or single-Trp/F339W NhaA grow only on the selective media at pH 7, the respective strains constructed on the wild-type background grow similar to the WT on all selective media (Table S1). Which of the native Trp residues is important for growth of the single-Trp/F136W or single-Trp/F339W mutants under extreme stress conditions is unknown at present.
Protein Expression and Antiport Activity in Membrane Vesicles.
Protein expression and the Na+/H+ or Li+/H+ antiport activity of Trp-less NhaA and its variants were measured in everted membrane vesicles isolated from EP432 cells transformed with plasmids encoding given variants. As compared to the control, expression of Trp-less NhaA was 15% and that of the variants 23%, 24%, and 18% for single Trp/F136W, single Trp/W258, and single Trp/F339W, respectively (Table S2). Because all variants are expressed from multicopy plasmids, the low level of expression is above the level expressed from a single chromosomal gene, which confers a Na+-resistant phenotype (15). Furthermore, the amount of protein expressed with each variant is sufficient for measuring Na+/H+ antiporter activity in everted membrane vesicles, as well as metal-affinity purification.
Na+/H+ antiporter activity of the NhaA variants was estimated from the change in ΔpH induced by either Na+ or Li+ as measured by acridine orange fluorescence, a qualitative probe of ΔpH (Fig. S1). Membrane vesicles isolated from EP432 cells transformed with plasmid pAXH3 (encoding WT NhaA) or the empty vector pBR322 served as positive or negative controls, respectively (Fig. S1 and Table S2). The apparent KM values for Na+ and Li+ at pH 8.5 and the extent of activity (maximal dequenching) at pH 8.5 were determined for each mutant (Table S2). Both Trp-less NhaA and its variant single Trp/F339W exhibit similar maximum dequenching (50–70%) and an apparent KM for both Na+ and Li+ about 10-fold higher than that of the WT (Fig. 1B and Table S2). In contrast, maximum dequenching of single Trp/F136W is 23–31%, and the apparent KM for Na+ and Li+ is very similar to WT. Variant single Trp/W258 has kinetic parameters that are very similar to WT (Fig. 1B and Table S2).
As shown previously (18), NhaA mutations that alter activation of NhaA at alkaline pH exhibit abnormal pH dependence even at saturating cation concentrations. The pH profile of the Trp-less and variants was not affected by changing Na+ concentration (10 or 100 mM; Fig. 1B). The profile for single Trp/W258 is identical to that of the WT, whereas those of the three other variants is slightly shifted to more alkaline pH.
Because all of the Trp-less variants grow on selective media at pH 7 at least, exhibit substantial antiport activity with both Na+ and Li+, and manifest a pH dependence only slightly different from WT, it is concluded that the structures are hardly impaired, if at all.
Trp Fluorescence of WT and Trp-Less NhaA.
Steady-state Trp fluorescence emission spectra were measured with given purified NhaA variants solubilized in DDM at pH 6.0 or 8.5 in the presence or absence of Li+ (or Na+). In most cases, excitation was at 295 nm to avoid excitation of the native Tyr residues in NhaA (19) (Fig. 2). Excitation at 290 nm gave higher emission intensities than expected, but very similar overall behavior.
The steady-state emission spectrum of WT NhaA at pH 8.5 exhibits a broad maximum centered at 335 nm (Fig. 2A), which is characteristic of Trp (19). In marked contrast, emission between 320 and 400 nm is essentially absent from the spectrum of Trp-less NhaA (Fig. 2A). The low residual emission can be ascribed to the eight Tyr residues in NhaA. A pH shift from 6 to 8.5 or addition of Li+ (Fig. 2A) (or Na+) has no effect on emission of Trp-less NhaA.
Single Trp/F136W, Effect of pH.
The steady-state emission spectrum of single Trp/F136W at pH 8.5 exhibits a typical Trp fluorescence spectrum with a broad maximum at around 343 nm (Fig. 2B). Remarkably, changing pH from pH 6.0 to 8.5 causes a red shift in the emission maximum and a marked increase in intensity (Fig. 2B). The fluorescence changes are not affected by addition of either 10–50 mM Li+ (or Na+) at either pH (Fig. 2B).
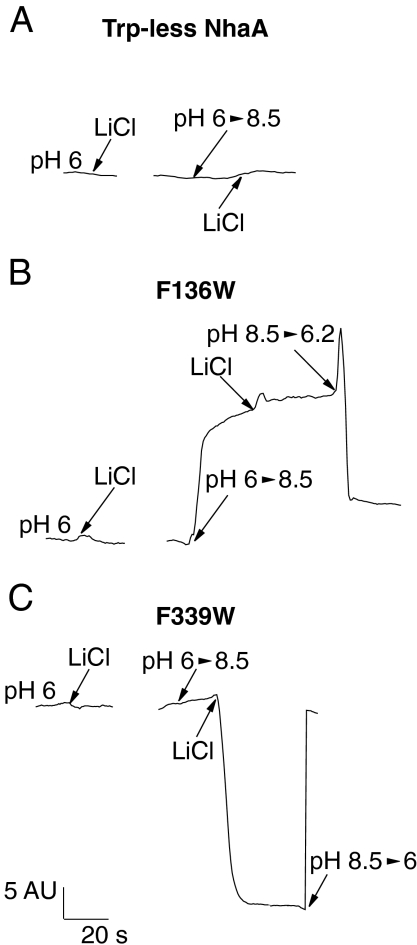
Fluorescence of single Trp/F136W was also measured in a time-dependent mode at 338 nm and compared to that of Trp-less NhaA (Fig. 3 A and B). As shown in Fig. 2A, neither a pH shift from pH 6.0 to 8.5 nor addition of Li+ (or Na+) has any effect on the fluorescence of Trp-less NhaA (Fig. 3A). In contrast, a shift in pH from 6.0 to 8.5 dramatically increases fluorescence of single Trp/F136W (Fig. 3B). A subsequent shift in pH from pH 8.5 to 6.2 reverses the effect, and fluorescence returns to a low level (Fig. 3B). Again, the presence of 10–50 mM LiCl had no effect (Fig. 3B).
Fig. 3.
Two different conformational changes, a pH induced and a Li+ induced, are reflected by the fluorescence changes of single Trp/F136W and Trp/F339W, respectively. The reaction mixtures were as in Fig. 2, and the fluorescence emission was measured with the time-dependent mode of the spectrofluorimeter. The excitation was at 295 nm and the emission was at the peak of each variant; 344, 335, and 338 nm for Trp-less NhaA, single Trp/F136W, and single Trp/F339W, respectively.
The pH profile for fluorescence of single Trp/F136W is shown in Fig. S2A. An increase in pH from 6.0 to 6.5 has no effect, but above pH 6.5, progressive increases in fluorescence are observed up to pH 8.0 with no further increase at pH 8.5. Taken together, the findings with single Trp/F136W reveal a reversible pH-induced conformational change of NhaA over the physiological pH range, which is independent of the presence of Li+ (or Na+).
Single-Trp/F339W, Effect of Li+ (or Na+).
The steady-state emission spectrum of single Trp/F339W exhibits a maximum at around 334 nm (Fig. 2C). In contrast to single-Trp/F136W fluorescence, a pH change between pH 6.0 and 8.5 in either direction with single Trp/339 has no effect on the emission spectrum (Figs. 2C and 3C). However, addition of either Li+ (or Na+) quenches fluorescence with little or no effect on λmax at pH 8.5 (Figs. 2C and 3C). Furthermore, no effect of Li+ (or Na+) is observed at pH 6.0 (Fig. 3C), indicating antiport activity is required for the effect of these cations. Li+-induced fluorescence quenching is fully reversed by shifting the pH back from pH 8.5 to 6.0 (Fig. 3C). In addition, between pH 6.0 and 7.0, Li+ has no effect on single-Trp/F339W fluorescence (Fig. S2B). But above pH 7, Li+ (or Na+)-induced quenching increases progressively up to pH 8.5. Hence, a pH change by itself does not affect the fluorescence of single Trp/F339W, but Li+ (or Na+) does so markedly in a pH-dependent manner.
Single-Trp/W258 NhaA.
The fluorescence emission spectrum of single Trp/W258 at pH 8.5 exhibits a maximum at around 337 nm (Fig. 2D) with a significantly increased λmax and a sharper maximum than those of single Trp/F136W or single Trp/F339W (compare Fig. 2D with Fig. 2 B and C). A shift from pH 6.0 to 8.5 leads to a small decrease in λmax, and addition of Li+ at pH 8.5 causes a decrease of similar magnitude (Fig. 2D). Furthermore, the pH profiles of the fluorescence changes at W258 are very similar to those observed for single Trp/F136W or F339W, and in addition (as shown below), when the lethal mutation D164N is also incorporated into each variant, the fluorescence changes are eliminated. Therefore, it is suggested that the fluorescence changes observed with single Trp/W258 reflect pH- and ligand-induced changes similar to those monitored with single Trp/F136W (Figs. 2B and 3B) and single Trp/F339W (Figs. 2C and 3C), respectively, but are of much lower magnitude.
Ligand-Induced Fluorescence Changes Require Functional Antiport Activity.
Importantly, the Na+/Li+ fluorescence changes in single Trp/F339W and probably single Trp/W258 are specific because neither choline chloride (50 mM) nor KCl (50 mM) has any effect on fluorescence emission (Fig. S3). The results strongly suggest that the Li+ (or Na+)-induced fluorescence changes reflect a conformational change induced by the binding of Li+/Na+ to NhaA or a conformational change following binding.
To test this idea further, the effect of Li+ was examined quantitatively with single Trp/F339W (Fig. S4), which exhibits a single exponential increase in fluorescence over a range of Li+ concentration from 0 to 100 mM. The K0.5 is 0.2 mM for Li+, which is similar to the apparent KM of 0.12 mM for Li+/H+ antiport activity in membrane vesicles containing single Trp/F339W (Table S2).
To determine whether the Trp fluorescence changes are related to the function of NhaA, the inactivating mutation D164N (20) was introduced into each single-Trp variant. Indeed, EP432 expressing single Trp/F136W/D164N, single Trp/F339W/D164N, or single Trp/W258/D164N grow on LBK (Luria broth with KCl instead of NaCl), but cannot grow on the salt-selective media (Table S1). The respective proteins (Fig. 2 E and F) exhibit Trp emission spectra similar to those of the corresponding proteins of the parent strains. However, 75% of the pH-induced fluorescence signal of single Trp/F136W was absent in single Trp/F136W/D164N (Fig. 2E), the large Li+-induced fluorescence signal was totally absent in single Trp/F339W/D164N (Fig. 2F), and single Trp/W258/D164N lost completely the Li+-induced signal and most of the pH-induced signal.
Discussion
Similar to other membrane proteins, NhaA contains multiple (eight) Trp residues located predominately at the membrane/water interface (Fig. 1A). When replaced with Phe (W37F, W126F, W216F, W309F, W360F) and Cys (W62C W268C), it is demonstrated here that none of the Trp residues in NhaA is essential for antiport activity (Table S1). Thus, EP432/Trp-less NhaA grows on all selective media, shows substantial Li+ (or Na+)/H+ antiport activity and a slightly alkaline-shifted pH dependence (Fig. 1B and Table S2). Hence, Trp-less NhaA provides a basis for introducing single-Trp residues at strategic sites as intrinsic fluorescence reporters (19) of NhaA conformational changes.
In contrast to Trp-less NhaA, which lacks a typical Trp fluorescence λmax at around 340 nm (19) (Fig. 2A), the three Trp-less NhaA variants, each with a single-Trp residue in the approximate middle of the membrane (Fig. 1A and ref. 9), exhibit characteristic Trp fluorescence spectra (Fig. 2).
Although the position of the λmax for either single Trp/F339W or single Trp/W258 is not significantly affected by pH or by the presence of Li+ or Na+, λmax for single Trp/F136W (338 nm at pH 6) is red shifted to 343 nm at pH 8.5 (Fig. 2B). Hence, the effect of pH on the fluorescence emission spectrum of single Trp/F136W is variant specific. Notably, over this pH range, the fluorescence of Trp in solution is unaffected by pH but is highly sensitive to solvent polarity. Thus, when embedded in a protein, Trp can report on the local environment and thereby monitor conformational changes and/or direct interaction with substrates (19). The red shift in the spectrum observed with single Trp/F136W at physiological pH most likely reflects a change in the local environment from a lower to a higher dielectric. Indeed, based on both experimental data (16) and computational analysis (21, 22), the cytoplasmic funnel (where F136W is located) is filled with water at physiological pH.
Strikingly, the fluorescence intensity of each single-Trp variant changes in a different manner when Li+ or Na+ concentration and/or pH are changed (Fig. 2 B–D). Possibly, the intensity changes may be related to conformational changes of the Trp residues or in residues in the vicinity of the Trp residues that cause quenching or energy transfer from the Trp to other amino acids (12). Although a precise mechanism for the fluorescence changes is unknown, the data presented show that they reflect conformational changes induced by Li+/Na+ and/or pH in NhaA.
Regulation of NhaA Na+/H+ antiport activity by pH is a most interesting characteristic that is shared by other eukaryotic and prokaryotic Na+/H+ antiporters (reviewed in ref. 10). NhaA is inactive below pH 6.5 and its rate increases by over three orders of magnitude between pH 7.0 and 8.5 (1, 6). Two scenarios, which are not mutually exclusive, have been advanced to explain activation at alkaline pH: (i) direct competition between H+ and Na+ or Li+ at a single cation-binding site that is alternatively exposed on either side of the membrane. Such competition has been documented recently for antiport (23) and is consistent with an alternating access mechanism (23–25). Accordingly, the conformational changes observed here with detergent solubilized NhaA reflect either ligand binding or a reaction following it. (ii) In addition to being a substrate for NhaA, H+ has a regulatory role and changes NhaA from an inactive to an active conformation that catalyzes antiport.
The pH-induced fluorescence changes observed with single Trp/F136W (Figs. 2B and 3B, and Fig. S2A) very likely reflect regulatory effects of pH on NhaA conformation that lead to activation at alkaline pH for the following reasons: (i) Increasing pH from pH 6.0 to 8.5 induces a red shift in λmax and a progressive, reversible increase in fluorescence of single Trp/F136W with pH (Figs. 2B and 3B). (ii) The profile for the pH-induced fluorescence changes is very similar to that of the pH activation of NhaA antiport (Fig. S2 A and C). (iii) Neither Na+ nor Li+ (50 mM) affects pH-induced fluorescence changes at any pH tested. Hence, Na+ or Li+ do not compete with the H+ for the pH-induced fluorescence changes observed with single Trp/F136W. (iv) pH-induced fluorescence changes are dependent on a functional NhaA molecule because single Trp/F136W/D164N (20), which is devoid of antiport activity, loses essentially all of the pH-induced fluorescence change (Fig. 2E). (v) Trp258, which is located in proximity to the pH sensor and remote from both Asp164 and Phe136 (distances between the respective α-carbons are 20 and 15 Å), appears to monitor the pH-induced conformational change as observed with single Trp/F136W, albeit much less dramatically. (vi) Importantly, as seen with single Trp/F136W, the pH-induced fluorescence change with W258 is reversible, requires functional NhaA, and shows pH dependence similar to that of single Trp/F136W (Fig. 2D).
Previous results strongly support the contention that a conformational change induced solely by increased pH activates NhaA (reviewed in ref. 3). Thus, pH-induced conformational changes independent of the presence of Na+/Li+ were experimentally identified at physiological pH. A Li+/Na+-independent pH-induced conformational change, with a pH profile very similar to that observed here with single Trp/F136W (Fig. S2A) has been identified recently in helix I by cryoelectron microscopy (26). Remarkably, the crystal structure shows that Phe136 is in close proximity to TM I, where the pH sensor is located (21). The distance between the Cα of Gly15 and Phe136 is approximately 5 Å (9). Finally, computational analysis of the NhaA structure suggests that Asp133 may be important for pH regulation (27).
Although neither Na+ nor Li+ are observed in the NhaA structure (9), the residues that most likely form the Na+-binding site are the most conserved [Asp164, Asp163 (TM V), Asp133, and Thr132 (TM IV)] and those shown to be essential (Asp164, Asp163) (9) (Fig. 1A). Mutations that affect cation translocation cluster at this location (3, 10). Neither Phe339 nor Trp258 (16) are components of the cation-binding site nor are they essential residues (Tables S1 and S2). Nevertheless, both exhibit Li+/Na+-induced fluorescence changes, yet the response of single Trp/F339W is at least five times greater, which may be due to the closer proximity of the Trp at position 339 to the binding site (Fig. 1A). Although widely separated, computational analysis (20) suggest tight functional coupling between the pH sensor and the binding site. The properties of the Li+/Na+-induced fluorescence changes with single Trp/F339W and single Trp/W258 indicate that both Trp residues reflect substrate binding to NhaA indirectly: (i) high specificity for Na+ and Li+—addition of either choline/Cl or KCl has no effect on fluorescence (Fig. S3). (ii) The effects on fluorescence are reversible (Fig. 3C). (iii) NhaA must be functional, because lethal mutations in the putative cation-binding site (single Trp/F339W/D164N and single Trp/W258/D164N) eliminate Li+- or Na+-induced fluorescence quenching (Fig. 2F). (iv) Dependence on cation concentration is shown in Fig. S4. (v) A pH profile very similar to that shown for pH activation of NhaA is observed with everted membrane vesicles or proteoliposomes reconstituted with purified proteins (Fig. S2 B and D). (vi) In strong support of this conclusion, cryoelectron microscopy shows directly that there is a ligand-induced conformational change in TM IVp between pH 7.0 and 8.5 (26), a pH range very similar to that used here with single Trp/F339W (Fig. S2 B) and single Trp/W258. Furthermore, the study also shows that TM IVp moves toward the active site and therefore toward F339W as well (Fig. 1A). A pH-dependent Li+/Na+-induced conformational change is also observed at the N-terminal portion of helix IX where W258 is located (23, 28).
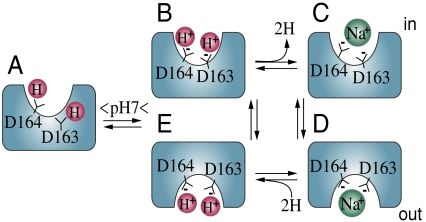
Why is a pH-dependent, Li+/Na+-induced conformational change observed with single Trp/F339W without a H+-induced conformational change? The X-ray structure of NhaA obtained from crystals prepared at pH 4.0 (2, 9) and results presented here provide the following working hypothesis (Fig. 4). In the X-ray structure, only Asp164, a major component of the cation-binding site, is exposed to the cytoplasmic funnel, whereas Asp163, another major component, is occluded (Fig. 4A; ref. 9). Furthermore, structure-based computation shows that Asp163, Asp164, and Asp133 in the binding site have a pKa above 11 (21) and therefore remain protonated at physiological pH unless a conformational change (most likely pH induced) exposes the binding site and reduces the pKa so that Li+/Na+ can now compete with the H+ and induce a conformational change directly related to catalytic activity. Taken together, by using site-directed Trp fluorescence, we revealed two conformational changes in NhaA: Step one revealed at F136W and W258— H+ binds to a site different from the active site and elicits a conformational change to activate NhaA by exposing the two aspartates (D163 and D164) to the cytoplasmic funnel and possibly also reducing their pKa so that they now ionize at physiological pH (Fig. 4 A and B). Step two monitored at F339W and W258—Li+/Na+ binds to the active site and releases the H+ from the active site (Fig. 4 C and D). Notably, because the crystal structure shows that the active site is within a delicately electrostatically balanced environment (9), entrance of Na+ or Li+ can also drastically change the pKa of the carboxyls. Following Na+/Li+ binding, translocation of the cation takes place according to the alternating access mechanism as has recently been shown (23) (Fig. 4 B–E).
Fig. 4.
Schematic working model of NhaA in the inactive, pH-activated, and active ion-translocating states. See text.
Notably, the identical pH dependence of the pH-induced and the ligand-induced conformational changes (Fig. 2 A and B) ensures that a pH signal, of a similar magnitude, elicits both conformational changes. Finally, we suggest that the conformational changes are likely sequential—the pH-induced conformational change activates NhaA and therefore precedes the ligand-induced conformational change which reflects the antiport activity of NhaA.
Materials and Methods
Bacterial Strains and Culture Conditions.
EP432 (melBLid, ΔnhaA1∷kan, ΔnhaB1∷cat, ΔlacZY, thr1) is an Escherichia coli K-12 derivative. TA16 is nhaA+nhaB+lacIQ (TA15lacIQ) and otherwise isogenic to EP432 (6). Cells were grown in modified L broth (LBK) (1). Antibiotics were 100 μg/mL ampicillin and/or 50 μg/mL kanamycin. To test the resistance to Li+ and Na+, EP432 cells transformed with the respective plasmids were grown on LBK to OD600 of 0.5. Samples (5 μL) of serial 10-fold dilutions of the cultures were spotted onto agar plates containing the indicated concentrations of NaCl or LiCl at the various pH values and incubated for 2 d.
Plasmids.
Plasmid pAXH (previously called pYG10), a pET20b (Novagen) derivative encodes His-tagged NhaA (29). A derivative of pAXH, pAXH2, lacks BglII site at position 3382 (30). A derivative of pAXH2, pAXH3, contains BstXI silent site at position 248 in nhaA. All plasmids carrying mutations are designated by the name of the plasmid followed by the mutation.
Mutagenesis.
Site-directed mutagenesis was conducted following a polymerase chain-reaction-based protocol (31). To construct Trp-less NhaA, Trp were stepwise replaced by Phe (Table S1). The mutants, pAXH3-W62C and pAXH3-W258C were constructed by insertion of EcoRI-BglII fragment of pCL-AXH3-W62C (15) and BstxI-XcmI fragment of pCL-AXH3-W258C (16) into pAXH3. The pAXH3-W62C-W258C was constructed by insertion of BglII -XcmI fragment of plasmid pAXH3-W258C into pAXH3-W62C. The plasmids p-single-Trp/F136W/D164N, p-single-Trp/W258/D164N, and p-single-Trp/F339W/D164N were constructed by introduction of D164N mutation into p-single-Trp/F136W, p-single-Trp/W258, and p-single-Trp/F339W. All mutations were verified by DNA sequencing of the entire gene, through the ligation junction with the vector plasmid.
Isolation of Membrane Vesicles, Assay of Na+/H+ Antiporter Activity.
EP432 cells transformed with the respective plasmids were grown in LBK and everted vesicles were prepared and used to determine the Na+/H+ or Li+/H+ antiporter activity using acridine orange (32).
Detection, Quantitation in the Membrane, and Purification of NhaA Variants.
Total membrane protein and the level of His-tagged NhaA variants in the membrane were determined as described (29, 33). NhaA protein variants were affinity purified (Ni2+-nitrilotriacetate-agarose, Qiagen) as previously described (16), but the protein was eluted in a buffer containing 300 mM imidazole (pH 7.9), 500 mM choline chloride, 20 mM Tris/Cl (pH 7.9), 0.015% n-DDM, and 10% sucrose, and dialyzed overnight at 4 °C in acid elution buffer (29) with 10% sucrose.
Fluorescence Measurements.
The reaction mixture (800 μL) contained very low Na+ (1–2 μM) as determined by atomic absorption, 20 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane, 150 mM choline chloride, 5 mM MgCl2, 10% sucrose, and 0.015% DDM, and incubated at 25 °C with magnetic stirring for 1 min. Then protein was added to yield 0.3–1.0 μM with or without other additions, incubation continued for 1 min, and the emission spectra were measured. For changing the pH in the reaction mixture, different amounts of 3.8 M Tris or 3.7% HCl were added.
Steady-state fluorescence was monitored in a Perkin Elmer LS45 Luminescence Spectrometer. Tryptophan emission spectra were recorded with an excitation wavelength of 290 or 295 nm as indicated. Time-dependent traces were recorded with excitation at 295 nm and emission wavelengths at 338–342 nm.
Supplementary Material
Acknowledgments.
E.P. thanks the European Drug Initiative on Channels and Transporters for Grant EU EP7 and the USA–Israel Binational Science Foundation for Grant BSF 20050130.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109256108/-/DCSupplemental.
References
- 1.Padan E, Bibi E, Ito M, Krulwich TA. Alkaline pH homeostasis in bacteria: New insights. Biochim Biophys Acta. 2005;1717:67–88. doi: 10.1016/j.bbamem.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padan E. The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem Sci. 2008;33:435–443. doi: 10.1016/j.tibs.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:C223–239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 5.Fliegel L. Molecular biology of the myocardial Na+/H+ exchanger. J Mol Cell Cardiol. 2008;44:228–237. doi: 10.1016/j.yjmcc.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 7.Taglicht D, Padan E, Schuldiner S. Proton-sodium stoichiometry of NhaA, an electrogenic antiporter from Escherichia coli. J Biol Chem. 1993;268:5382–5387. [PubMed] [Google Scholar]
- 8.Orlowski J, Grinstein S. Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr Opin Cell Biol. 2007;19:483–492. doi: 10.1016/j.ceb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 10.Padan E, Kozachkov L, Herz K, Rimon A. NhaA crystal structure: Functional-structural insights. J Exp Biol. 2009;212:1593–1603. doi: 10.1242/jeb.026708. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y, et al. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature. 2011;474:109–113. doi: 10.1038/nature09971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnova I, Kasho V, Sugihara J, Kaback HR. Probing of the rates of alternating access in LacY with Trp fluorescence. Proc Natl Acad Sci USA. 2009;106:21561–21566. doi: 10.1073/pnas.0911434106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menezes ME, Roepe PD, Kaback HR. Design of a membrane transport protein for fluorescence spectroscopy. Proc Natl Acad Sci USA. 1990;87:1638–1642. doi: 10.1073/pnas.87.5.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vazquez-Ibar JL, Guan L, Svrakic M, Kaback HR. Exploiting luminescence spectroscopy to elucidate the interaction between sugar and a tryptophan residue in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 2003;100:12706–12711. doi: 10.1073/pnas.1835645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herz K, Rimon A, Olkhova E, Kozachkov L, Padan E. Transmembrane segment II of NhaA Na+/H+ antiporter lines the cation passage, and Asp65 is critical for pH activation of the antiporter. J Biol Chem. 2010;285:2211–2220. doi: 10.1074/jbc.M109.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzubery T, Rimon A, Padan E. Structure-based functional study reveals multiple roles of TMS IX and loop VIII-IX in NhaA Na+/H+ antiporter of Escherichia coli at physiological pH. J Biol Chem. 2008;283:15975–15987. doi: 10.1074/jbc.M800482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimon A, Tzubery T, Padan E. Monomers of NhaA Na+/H+ antiporter of Escherichia coli are fully functional yet dimers are beneficial under extreme stress conditions at alkaline pH in the presence of Na+ or Li+ J Biol Chem. 2007;282:26810–26821. doi: 10.1074/jbc.M704469200. [DOI] [PubMed] [Google Scholar]
- 18.Galili L, Herz K, Dym O, Padan E. Unraveling functional and structural interactions between transmembrane domains IV and XI of NhaA Na+/H+ antiporter of Escherichia coli. J Biol Chem. 2004;279:23104–23113. doi: 10.1074/jbc.M400288200. [DOI] [PubMed] [Google Scholar]
- 19.Lackowicz JR. Principles of Fluorescence Spectroscopy. New York: Plenum Publishers; 1999. [Google Scholar]
- 20.Olkhova E, Kozachkov L, Padan E, Michel H. Combined computational and biochemical study reveals the importance of electrostatic interactions between the “pH sensor” and the cation binding site of the sodium/proton antiporter NhaA of Escherichia coli. Proteins. 2009;76:548–559. doi: 10.1002/prot.22368. [DOI] [PubMed] [Google Scholar]
- 21.Olkhova E, Hunte C, Screpanti E, Padan E, Michel H. Multiconformation continuum electrostatics analysis of the NhaA Na+/H+ antiporter of Escherichia coli with functional implications. Proc Natl Acad Sci USA. 2006;103:2629–2634. doi: 10.1073/pnas.0510914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olkhova E, Padan E, Michel H. The influence of protonation states on the dynamics of the NhaA antiporter from Escherichia coli. Biophys J. 2007;92:3784–3791. doi: 10.1529/biophysj.106.098269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mager T, Rimon A, Padan E, Fendler K. Transport mechanism and pH regulation of the Na+/H+ antiporter NhaA from Escherichia coli: An electrophysiological study. J Biol Chem. 2011;286:23570–23681. doi: 10.1074/jbc.M111.230235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 25.Kaback HR, Smirnova I, Kasho V, Nie Y, Zhou Y. The alternating access transport mechanism in LacY. J Membr Biol. 2010;239:85–93. doi: 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Appel M, Hizlan D, Vinothkumar KR, Ziegler C, Kuhlbrandt W. Conformations of NhaA, the Na+/H+ exchanger from Escherichia coli, in the pH-activated and ion-translocating states. J Mol Biol. 2009;388:659–672. doi: 10.1016/j.jmb.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Arkin IT, et al. Mechanism of Na+/H+ antiporting. Science. 2007;317:799–803. doi: 10.1126/science.1142824. [DOI] [PubMed] [Google Scholar]
- 28.Tzubery T, Rimon A, Padan E. Mutation E252C increases drastically the Km value for Na+ and causes an alkaline shift of the pH dependence of NhaA Na+/H+ antiporter of Escherichia coli. J Biol Chem. 2003;279:3265–3272. doi: 10.1074/jbc.M309021200. [DOI] [PubMed] [Google Scholar]
- 29.Olami Y, Rimon A, Gerchman Y, Rothman A, Padan E. Histidine 225, a residue of the NhaA-Na+/H+ antiporter of Escherichia coli is exposed and faces the cell exterior. J Biol Chem. 1997;272:1761–1768. doi: 10.1074/jbc.272.3.1761. [DOI] [PubMed] [Google Scholar]
- 30.Galili L, Rothman A, Kozachkov L, Rimon A, Padan E. Trans membrane domain IV is involved in ion transport activity and pH regulation of the NhaA-Na+/H+ antiporter of Escherichia coli. Biochemistry. 2002;41:609–617. doi: 10.1021/bi011655v. [DOI] [PubMed] [Google Scholar]
- 31.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 32.Rosen BP. Ion extrusion systems in Escherichia coli. Methods Enzymol. 1986;125:328–336. doi: 10.1016/s0076-6879(86)25028-4. [DOI] [PubMed] [Google Scholar]
- 33.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.