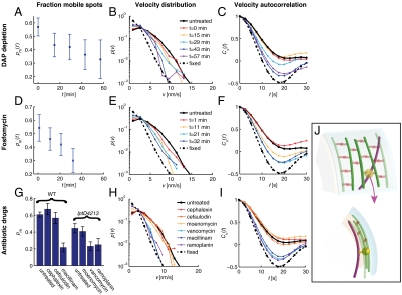
Fig. 3.
MreB rotation requires peptidoglycan substrate availability and the activity of transglycosylases and transpeptidases. For each dataset, we determined the fraction of mobile MreB spots pm(t) (A, D, and G), the distribution of the velocities of all MreB spots p(v) (B, E, and H), and the velocity autocorrelation function Cv(t) (C, F, and I). Nonmutant cells (solid black line) and chemically fixed cells (dashed line) serve as references in the latter two analyses. For A–C, asd-1 mutant cells were depleted of DAP at time zero and analyzed at the subsequent indicated time points. For D–F, cells were treated with the MurA inhibitor fosfomycin at time zero and analyzed at subsequent indicated time points. For G–I, nonmutant (WT) and lptD4213 mutant cells were analyzed between 10–20 min after treatment with the indicated antibiotic drug. (J) Top (Upper) and side (Lower) views of a model for cell-wall-synthase-driven MreB rotation. Here, we propose that the transmembrane cell-wall synthesis complex (yellow) acts in the periplasm (between the cyan outer membrane and peach inner membrane) to polymerize glycan strands (green) and form peptide cross-links (orange) to incorporate the strands into the existing peptidoglycan network, thereby causing the associated MreB filaments (magenta) to rotate in the cytoplasm.