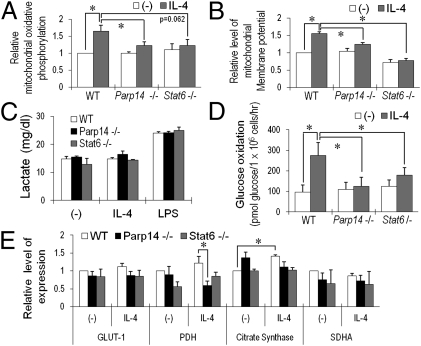
Fig. 4.
PARP14 mediates IL-4–enhanced mitochondrial activity in B cells. (A) As detailed in Materials and Methods, mitochondrial oxidative phosphorylation was measured in B cells (WT, Parp14-, and Stat6-null) using a flow cytometric assay after culture (20 h) in medium ± IL-4. Specific mean fluorescent intensities (MFI) of mitochondrial signals for B220+ cells in matched samples of each independent experiment were normalized to the MFI of the WT sample cultured without added cytokine. Bar graphs represent the means (±SEM) of normalized values from these independent replicates. (B) PARP14 dependence of the IL-4–enhanced mitochondrial membrane potential in B cells. Mitochondrial membrane potential in samples as in A was measured using DiOC6 (3) dye; shown are mean (±SEM) data from independent replicate experiments. Signals for B220+ cells in matched samples of each independent experiment were normalized as in A. Bar graphs represent the means (±SEM) of three independent replicates. (C) Lactate in the medium was measured for purified B cells (WT, Parp14−/−, and Stat6−/−) after culture (20 h) in the presence or absence of IL-4 (5 ng/mL) or LPS (1 μg/mL). (D) PARP14- and Stat6-dependent enhancement of glucose oxidation by IL-4. Using B cells as in glycolysis assays, glucose oxidation was assayed as detailed in Materials and Methods. (E) PARP14 impacts PDH gene expression in IL-4–treated B cells. After first determining an optimal time for measuring induction by IL-4, RNA was isolated from B cells cultured for 4 h with and without IL-4; levels of mRNAs encoding metabolic genes were analyzed by quantitative real-time RT-PCR. Bar graphs show mean (±SEM) data from three independent experiments, with mRNA levels in each experiment normalized to WT B cells without IL-4 treatment and then averaged, as in A (*P < 0.05).