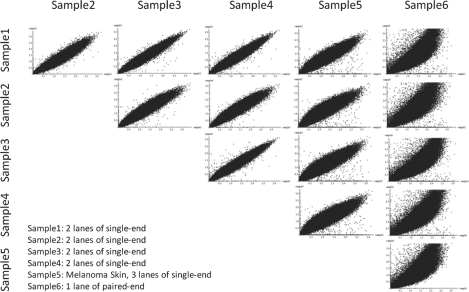
Fig. 2.
Correlation of depth-of-coverage across exome sequencing samples. To demonstrate the consistency of capture and sequencing efficiency of individual exons represented by the depth-of-coverage per exon, the normalized individual exon coverage in all pairs of six-independent exomes were plotted. All 6 samples were captured using the Agilent SureSelect Human All Exon G3362. Samples 1–5 had mean base coverages of 36~39× as a result of 2 (Samples 1–4) or 3 (Sample 5) lanes of GAIIx single-end sequencing per sample. Sample 6 had mean base coverage of 60× as a result of 1 lane of GAIIx paired-end sequencing and demonstrates substantially different biases in individual exonic depth-of-coverage.