Abstract
Background
Pyoderma gangrenosum–like ulcers and cellulitis of the lower extremities associated with recurrent fevers in patients with X-linked (Bruton) agammaglobulinemia have been reported to be caused by Helicobacter bilis (formerly classified as Flexispira rappini and then Helicobacter strain flexispira taxon 8). Consistent themes in these reports are the difficulty in recovering this organism in blood and wound cultures and in maintaining isolates in vitro. We confirmed the presence of this organism in a patient’s culture by using a novel application of gene amplification polymerase chain reaction and electrospray ionization time-of-flight mass spectrometry.
Observation
An adolescent boy with X-linked agammaglobulinemia presented with indurated plaques and a chronic leg ulcer whose origin was strongly suspected to be an H bilis organism. Histologic analysis demonstrated positive Warthin-Starry staining of curvilinear rods, which grew in culture but failed to grow when sub-cultured. They could not be identified by conventional techniques. A combination of gene amplification by polymerase chain reaction and electrospray ionization time-of-flight mass spectrometry confirmed the identity of this organism.
Conclusions
This novel technology was useful in the identification of a difficult-to-grow Helicobacter organism, the cause of pyoderma gangrenosum–like leg ulcers in patients with X-linked agammaglobulinemia. Correct identification of this organism as the cause of pyoderma gangrenosum–like ulcers in patients with X-linked agammaglobulinemia is of great importance for the early initiation of appropriate and curative antibiotic therapy.
X-Linked Agammaglobulinemia (XLA), also known as Bruton agammaglobulinemia, is a primary immunodeficiency first described by Bruton1 in 1952 in an 8-year-old boy who had recurrent pneumococcal sepsis associated with a complete lack of γ-globulin and antibodies. Bruton found that this child was unable to make antibodies to pneumococcal and typhoid vaccines and that these infections were controlled by subcutaneous injections of γ-globulin. Subsequently, the defect in antibody production was shown to be caused by the absence or severe lack of mature B cells and plasma cells and was eventually linked to a defective BTK gene in 1993.2 Patients with XLA were found to be susceptible to infections by encapsulated organisms, such as Haemophilus influenzae and Streptococcus pneumoniae, as well as enteroviruses and parasites, such as Giardia lamblia. In a US registry of 201 patients,3 70% developed otitis media, 59% had sinusitis, and 62% had pneumonias. Enteroviral infections with coxsackievirus and echovirus inevitably led to meningoencephalitis and accounted for 35% of deaths among patients in the registry. Skin infections that encompassed pyoderma, cellulitis, and abscess occurred in 18% of registered patients.
REPORT OF A CASE
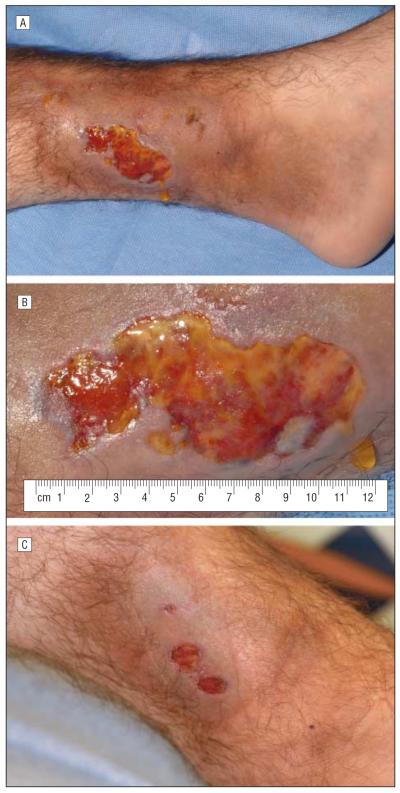
A 17-year-old boy had been diagnosed as having XLA at 6 months of age. He had presented initially with an ear infection at 2 months of age followed 4 months later by spontaneous gluteal ulcers. At 12 years of age he had developed a lump on the lateral aspect of the right leg after twisting his ankle. This lump eventually turned into a fibrotic and hyperpigmented plaque. At 15 years of age he had developed cellulitis of the right arm and since then has had multiple episodes of painless, oval lumps developing on his upper and lower extremities and the lateral aspect of the thorax. Several of these lesions were biopsied, but only inflammation was noted. Six months before admission, a scar on his left leg, that had resulted from a go-kart injury 3 years previously, turned lumpy; it ulcerated 2 months later. A 10-day course of amoxicillin clavulanate and topical ulcer care were not helpful. He was admitted to the Clinical Center of the National Institutes of Health for evaluation and treatment of the chronic ulcer. On admission, he was a well-developed, afebrile boy whose abnormal findings were confined to his legs. The skin over the distal third of both legs was hyperpigmented and markedly indurated. There was an irregularly shaped, 5.5 × 3.4-cm ulcer with surrounding erythema on the medial aspect of the left leg, proximal to the medial malleolus. The ulcer base was yellowish with an adherent serofibrinous film, especially along the periphery. The inner margins of the ulcer were somewhat rolled (Figure 1A and B). Skin punch biopsy specimens were taken from the ulcer edge and sent for histologic and microbiological studies.
Figure 1.
Ulcer on the leg of a 17-year-old boy. A, Leg ulcer with surrounding hyperpigmentation and induration at presentation. B, Same leg ulcer (close-up). C, Nearly healed leg ulcer 2½ months after treatment with systemic antibiotics.
RESULTS
A bone scan demonstrated soft-tissue inflammation, and magnetic resonance imaging revealed no evidence of osteomyelitis. The IgA and IgM levels were below normal, whereas the IgG level was in the lower range of normal. He had been receiving intravenous immunoglobulin every 3 weeks since 6 months of age. The rest of the routine laboratory test results were within normal limits. Blood culture was negative for bacteria. The edge of the ulcer was biopsied and sent for histopathologic and microbiological cultures with specific instructions to look for Flexispira and Helicobacter. Routine histologic analysis revealed a suppurative granulomatous reaction. Periodic acid–Schiff, Fite, Brown, Bren, and Gomori methenamine-silver stains were negative for bacteria and fungi. A Warthin-Starry silver stain of the skin biopsy specimen was positive for numerous curvilinear, rodlike organisms (Figure 2). The initial wound cultures yielded Streptococcus agalactiae.
Figure 2.
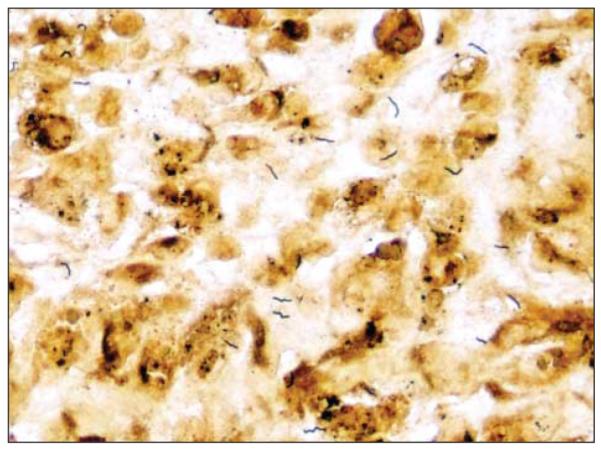
Curvilinear rods on Warthin-Starry staining of a biopsy specimen from the ulcer edge (original magnification ×400).
A team at the National Institutes of Health reported 2 patients with XLA who had bacteremia, skin infections, and bone infections caused by a Flexispira/Heliobacter organism.4,5 Because one of us (M.L.T.) had participated in the evaluation of 1 of these patients, we were certain that our patient was similarly infected. While awaiting specific microbiological results, he was sent home with intravenous tobramycin (140 mg every 8 hours) and meropenem (1 g every 8 hours). On follow-up 2½ months later, the ulcer was almost completely healed (Figure 1C), he reported feeling better, and he had gained 6.8 kg.
MICROBIOLOGICAL STUDIES
A total of 4 blood cultures were collected: 2 routine blood cultures were held for 7 days, and 2 were processed specifically for H bilis (incubated for 14 days; blind acridine orange stains and subcultures performed at 7 and 14 days). All blood cultures were negative. Two cultures collected from skin biopsy specimens of the ulcer edge were processed with each inoculated onto enriched blood agar and chocolate agar plates.
The plates were incubated in a microaerophilic atmosphere with supplemented hydrogen and carbon dioxide. After 24 hours of incubation, a few colonies of group B Streptococcus (S agalactiae) were observed on the media. Because organisms consistent with H bilis were observed in the Warthin-Starry–stained skin biopsy sample, the cultures were incubated for 7 days. On day 6, a thin film of growth was observed surrounding the colonies of S agalactiae. Gram stain and acridine orange stains of this growth revealed thin, curved rods; however, all subcultures of this growth were nonviable. Definitive identification of the organism by conventional gene sequencing could not be made because multiple organisms were present in the culture.
SPECIAL STUDIES
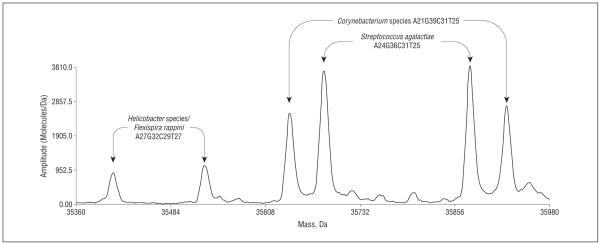
Ecker et al6 and Hofstadler et al7 have described the use of a panel of broad bacterial primer pairs targeting several ribosomal and housekeeping genes for unbiased amplification of known and unknown bacteria using a rapid microbial detection technique. Commercially known as the Ibis T5000 Biosensor System (Abbott Laboratories, Abbott Park, Illinois), this system uses polymerase chain reaction amplification of selected gene targets to produce a panel of amplified products, followed by separation of the products by their mass, using electron spray ionization time-of-flight mass spectrometry. Each amplified gene product, or amplicon, has a characteristic mass or molecular weight that is then compared with a database to determine the organism’s identity. Because the amplicons are separated by their mass, the identification of multiple species of bacteria in a clinical specimen or in vitro culture is possible. Analysis of our patient’s positive wound culture was performed by using a sterile needle to pick 10 discrete colonies. Each colony was resuspended in 500 μL of sterile water in a microfuge tube. The tube was heated for 5 minutes at 95°C and then centrifuged to settle cellular debris. Supernatant containing the genomic material was added to polymerase chain reaction tubes and amplified, and the amplicon products were separated and analyzed by the T5000 Biosensor System. The profile of 1 targeted gene is demonstrated in Figure 3. A mixture of 3 bacteria was present: S agalactiae (group B Streptococcus), a Corynebacterium that was recognized as a contaminant introduced during the handling of the culture plate, and a Helicobacter species. The Helicobacter isolate could only be identified at the genus level because multiple Helicobacter species had base compositions consistent with this detection (H bilis, Helicobacter canis, and Helicobacter trogontum). Detection of Helicobacter is consistent with the clinical history and histopathologic and microbiological findings.
Figure 3.
Spectral display showing 3 different bacterial detections from colonies on a primary culture plate: Streptococcus agalactiae, Corynebacterium species, and Helicobacter species. These are spectra from a primer pair that targets 16S rDNA. The paired peaks correspond to the sense and antisense strands of the polymerase chain reaction amplicon that are separated under conditions of ionization. The peaks are labeled with base composition of the amplicons and the organism that matches the composition.
COMMENT
The presentation of a boy with XLA and a chronic, pyoderma gangrenosum (PG)–like leg ulcer recalled a similar article4 for which one of the authors (M.L.T.) had consulted. In one of the cases discussed, biopsy specimens from the ulcer edge did not yield any organisms, but blood cultures and samples from scrapings of osteomyelitic bones yielded a fastidious, curved, gram-negative rod, Flexispira rappini, which was subsequently reclassified as H bilis. The ulcers healed after 9 months of intravenous antibiotics. That same article included another patient with XLA7 whose case had previously been reported in greater detail.5 He had had recurrent bouts of antibiotic-resistant sepsis accompanied by cellulitis of the right leg, from whose blood a Flexispira-like organism was cultured. Since then, there have been other reports of infection by Helicobacter species in patients with XLA. In 1 case, bacteria were visualized by acridine orange staining of pus from skin abscesses, but these could not be cultured. Bacterial DNA was directly extracted from the pus, and 16S ribosomal RNA gene sequence analysis identified a unique Helicobacter species closely related to F rappini and H bilis.8 By 16S ribosomal RNA sequencing of bacteria from blood cultures, F rappini was identified as the organism that caused fevers and cellulitis of the legs in another patient with XLA.9
As originally described by Brunsting et al,10 typical PG manifests as rapidly progressive, painful, suppurative ulcers with undermined, ragged, purplish borders. To quote directly from a major textbook in dermatology, “Seventy years later, the etiology of pyoderma gangrenosum is still unknown, but in all likelihood bacteria do not directly cause the disease, nor is it infectious in nature.”11 Gene sequencing techniques have recently been used to study 3 cases of postoperative PG. In these instances, the inability to detect microbial 16S ribosomal RNA resulted in earlier institution of systemic steroids, which promoted rapid healing of the ulcers.12
Over time, PG has come to be thought of as a reactive phenomenon secondary to a long list of diverse primary diseases, such as hematologic malignant neoplasms, rheumatoid arthritis, or inflammatory bowel disease. In fact, this was the explanation given for the 2 cases of PG that were encountered in a series of 15 cases of XLA reported from the Netherlands.13
This current report demonstrates that the large, PG-like ulcer in the lower extremity of our patient with XLA is an infectious ulcer caused by H bilis. Recognition of the infectious nature of PG-like ulcers in patients with XLA will forestall the use of high-dose systemic steroids, the traditional treatment for PG, which could cause infection to spread. Obtaining a definitive identification of this fastidious and difficult-to-culture organism from PG-like ulcers in patients with XLA is also important because appropriate antibiotic treatment is expensive and lasts for many months. Although traditional gene sequencing can be used to identify an isolated organism, the technology described in this report, electron spray ionization time-of-flight mass spectrometry, can be used to identify multiple species of bacteria that may be present in a culture or clinical specimen, such as those that occurred in this case.
Acknowledgments
Funding Support: This study was supported in part by protocol 06-I-0049 of the National Institute of Allergy and Infectious Diseases and Ibis Biosciences (Abbott Laboratories), whose employees performed the spectrometric analysis of polymerase chain reaction products using the Ibis T5000 Biosensor System.
Role of the Sponsors: The sponsors had no role in the design and conduct of the study; in the collection, analysis, and interpretation of data; or in the preparation, review, or approval of the manuscript.
Footnotes
Author Contributions: Drs Turner, Murray, and Uzel had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Turner, Murray, Sampath, Blyn, Ecker, Ranken, and Ivy. Acquisition of data: Turner, Murray, Jain, Uzel, Sampath, Blyn, Ecker, Ranken, Ivy, and Lee. Analysis and interpretation of data: Turner, Murray, Sampath, Blyn, Ecker, and Ranken. Drafting of the manuscript: Turner, Murray, Sampath, Blyn, Ecker, Ranken, and Ivy. Critical revision of the manuscript for important intellectual content: Turner, Murray, Jain, Uzel, Sampath, Blyn, Ecker, Ranken, and Lee. Administrative, technical, and material support: Turner, Murray, Jain, Uzel, Sampath, Blyn, Ecker, Ranken, Ivy, and Lee. Study supervision: Turner, Murray, Sampath, Blyn, Ecker, and Ranken.
Financial Disclosure: None reported.
REFERENCES
- 1.Bruton OC. Agammaglobulinemia. Pediatrics. 1952;9(6):722–728. [PubMed] [Google Scholar]
- 2.Tsukada S, Saffran DC, Rawlings DJ, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72(2):279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 3.Winkelstein JA, Marino MC, Lederman HM, et al. X-linked agammaglobulinemia: report and United States repository of 201 patients. Medicine (Baltimore) 2006;85(4):193–202. doi: 10.1097/01.md.0000229482.27398.ad. [DOI] [PubMed] [Google Scholar]
- 4.Cuccherini B, Chua K, Gill V, et al. Bacteremia and skin/bone infections in two patients with X-linked agammaglobulinemia caused by an unusual organism related to Flexispira/Helicobacter species. Clin Immunol. 2000;97(2):121–129. doi: 10.1006/clim.2000.4932. [DOI] [PubMed] [Google Scholar]
- 5.Weir S, Cuccherini B, Whitney AM, et al. Recurrent bacteremia caused by a “Flexispira”-like organism in a patient with X-linked (Bruton’s) agammaglobulinemia. J Clin Microbiol. 1999;37(8):2439–2445. doi: 10.1128/jcm.37.8.2439-2445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ecker DJ, Sampath R, Blyn LB, et al. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc Natl Acad Sci U S A. 2005;102(22):8012–8017. doi: 10.1073/pnas.0409920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofstadler SA, Sampath R, Blyn LB, et al. TIGER: the universal biosensor. Int J Mass Spectrom. 2005;242:23–41. [Google Scholar]
- 8.Han S-R, Schindel C, Genitsariotis R, Märker-Hermann E, Bhakdi S, Maeurer M. Identification of a unique Helicobacter species by 16S rRNA gene analysis in an abdominal abscess from a patient with X-linked hypogammaglobulinemia. J Clin Microbiol. 2000;38(7):2740–2742. doi: 10.1128/jcm.38.7.2740-2742.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerrard J, Alfredson D, Smith I. Recurrent bacteremia and multifocal lower limb cellulitis due to Helicobacter-like organisms in a patient with X-linked hypogammaglobulinemia. Clin Infect Dis. 2001;33(10):E116–E118. doi: 10.1086/323405. [DOI] [PubMed] [Google Scholar]
- 10.Brunsting LA, Goeckerman WH, O’Leary PA. Pyoderma (ecthyma) gangrenosum: clinical and experimental observations in 5 cases occurring in adults. Arch Derm Syphilol. 1930;22:655–680. [Google Scholar]
- 11.Wolff K, Stingl G. Pyoderma gangrenosum. In: Freedberg IM, Eisen AZ, Wolff K, Austen KF, Goldsmith LA, Katz SA, editors. Fitzpatrick’s Dermatology in General Medicine. 6th ed McGraw-Hill; New York, NY: 2003. p. 969. [Google Scholar]
- 12.Ferrandiz-Pulido C, Bartralot R, Fuente MJ, et al. Postoperative pyoderma gangrenosum: diagnostic value of 16s ribosomal RNA sequencing and review of the literature. Clin Exp Dermatol. 2009;34(5):598–602. doi: 10.1111/j.1365-2230.2008.03049.x. [DOI] [PubMed] [Google Scholar]
- 13.Van Der Hilst BW, Smits BW, Van Der Meer JWM. Hypogammaglobulinaemia: cumulative experience in 49 patients in a tertiary care institution. Neth J Med. 2002;60(3):140–147. [PubMed] [Google Scholar]