Figure 1.
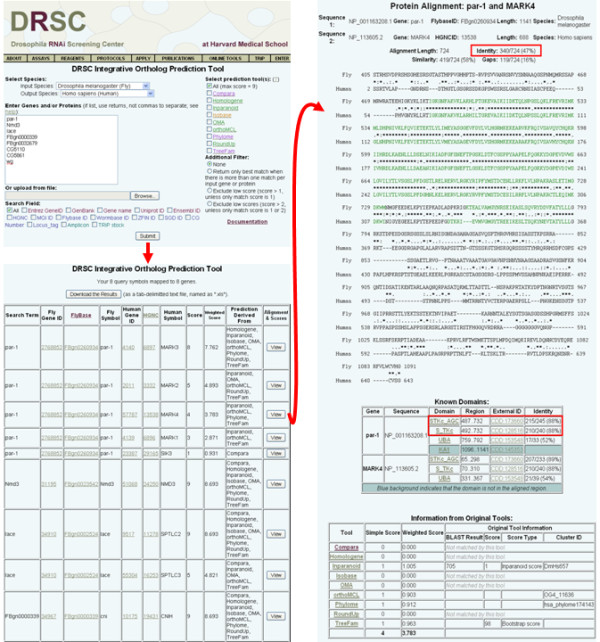
DIOPT facilitates identification of predicted orthologs based on various approaches, with flexible inputs and outputs. The DIOPT graphical user interface facilitates input of small or large gene or protein lists and is compatible with one or more of a variety of identifiers. Three filters are available: two of them exclude relationships that are predicted by < 2 or < 3 tools unless the only match score is equal to or lower than the threshold. The third one and most stringent filter limits outputs to the best-matching ortholog(s) per gene, as judged by the number of tools supporting the prediction. DIOPT provides a score based on the number of tools supporting the prediction and a weighted score based on the average of GO semantic similarity of orthologous pairs predicted by each tool using high quality GO functional annotation. In addition, DIOPT also provides the option to view the original score of each tool and a protein-protein alignment based on an updated proteome annotation (RefSeq release 44). Percent protein identity is calculated for both the overall alignment and domain regions.
