Abstract
We have recently identified apurinic/apyrimidinic endonuclease 1 (APE1) as an endoribonuclease that cleaves c-myc mRNA in vitro and regulates c-myc mRNA levels and half-life in cells. This study was undertaken to further unravel the RNA-cleaving properties of APE1. Here, we show that APE1 cleaves RNA in the absence of divalent metal ions and, at 2 mM, Zn2+, Ni2+, Cu2+, or Co2+ inhibited the endoribonuclease activity of APE1. APE1 is able to cleave CD44 mRNA, microRNAs (miR-21, miR-10b), and three RNA components of SARS-corona virus (orf1b, orf3, spike) suggesting that, when challenged, it can cleave any RNAs in vitro. APE1 does not cleave strong doublestranded regions of RNA and it has a strong preference for 3’ of pyrimidine, especially towards UA, CA, and UG sites at single-stranded or weakly paired regions. It also cleaves RNA weakly at UC, CU, AC, and AU sites in single-stranded or weakly paired regions. Finally, we found that APE1 can reduce the ability of the Dicer enzyme to process premiRNAs in vitro. Overall, this study has revealed some previously unknown biochemical properties of APE1 which has implications for its role in vivo.
Keywords: Endoribonuclease, APE1, CD44, miRNA, RNA structure
Introduction
Apurinic/apyrimidinic (AP) endonuclease 1 (APE1) is a 35 kDa protein with multifunctional enzymatic activities which include 3’ -5’ DNA exonuclease [1], 3’ phosphodiesterase [2], RNase H-like [3] and AP DNA endonuclease activities [4]. The AP DNA endonuclease function involves incision of the phosphodiester bond immediately 5’ to an AP site, generating products with 3’ hydroxyls and 5’ -deoxyribose phosphate termini [5]. The products generated are then processed by enzymes involved in the subsequent steps of the base excision repair (BER) pathway to fill the nucleotide gap and seal the nick in the DNA to complete the DNA AP site repair. In mammalian cells, APE1 is the major protein involved in initiating removal of abasic sites in DNA [6]. In addition to its DNA repair activity, the mammalian APE1 was also independently discovered to possess redox activity, hence it is also known as redox effector factor 1 (Ref-1) [7]. Such function of APE1 has been shown to keep transcription factors such as c-Jun and p53 in the reduced and active form [8,9].
Stemming from an interest in understanding the general role of endoribonucleases in the control of gene expression, and in finding the enzyme that endonucleolytically cleaves and regulates c-myc mRNA, we continued our efforts in the biochemical purification and identification of the responsible endoribonuclease [10]. Studies in cell lines [11,12], cell-free mRNA decay assay involving polysome extracts [13] as well as animals [14], have confirmed that the coding region of c-myc mRNA which encompasses a specific region termed the coding region determinant (CRD) is involved in the regulation of c-myc mRNA stability. Using c-myc CRD RNA as substrate, we unexpectedly identified APE1 as an enzyme with endoribonuclease activity [10]. We showed that APE1 preferentially cleaves c-myc CRD RNA at UA, UG and CA sites [10]. Using a specific siRNA against APE1 mRNA, we further showed that APE1 is in fact involved in the regulation c-myc mRNA levels and half-life in cells [10].
Given the demonstrated significance of APE1 in controlling c-myc mRNA and possibly other mRNAs in cells, it is thus warranted to further understand the biochemical RNA-cleaving properties of APE1. This study was undertaken to address the following questions. Can APE1 cleave other RNA molecules especially those that are implicated in diseases? If it does cleave other RNA molecules, does it still preferentially cleave at UA, UG, and CA dinucleotides at single-stranded regions only? What are the divalent metal ion requirements for the RNA-cleaving activity of APE1?
Materials and methods
Purification of recombinant human APE1
The plasmid pET15b-hAPE1, kindly provided by Dr. Sankar Mitra, containing the human APE1 cDNA was used to express recombinant APE1 in E.coli strain BL21(DE3) cells. The His-tagged APE1 was first purified using Ni-NTA column chromatography as previously described [10]. Following removal of the His-tag with thrombin, the recombinant protein was further purified to homogeneity by ion exchange chromatography as previously described [10]. Just prior to use, the recombinant protein was dialyzed for 5 h against 10 mM Tris-HCI, pH 7.4, 2 mM DTT, 2 mM magnesium acetate, and 50 mM potassium acetate, with two buffer changes. For use in divalent metal ion requirement experiments, the protein was dialyzed in Mg2+-free buffer containing 10 mM EDTA.
Construction of plasmids and DNA templates
Plasmids pUC19-Spike, pUC19-orf3, and pUC19 -orf1b were constructed using molecular subcloning techniques according to established protocols [15]. cDNA clones of SARS-coronavirus were gifts from Dr. Marco Marra (Genome Science Centre, BC Cancer Agency) and were used as template to amplify segments of cDNA of spike (nts 21482-21549), orf3 (nts 25260-25339), and orf1b (nts 14440-14508) [16]. The PCR primers used were: Spike forward primer, 5’ -CTC GGA TCC TAA TAC GAC TCA CTA TAG GCT AAA CGA ACA TGT TTA T-3’, Spike reverse primer, 5’ -CTC GAA TTC TGC ACC GGT CAA GGT CAC-3’ ; orf3 forward primer, 5’ -CTC GGA TCC TAA TAC GAC TCA CTA TAG GCG AAC TTA TGG ATT TGT T-3’, and orf3 reverse primer, 5’ -CTC GAA TTC GAG AAG CAT TGT CAA TTT-3’ ; orf1b forward primer, 5’ -CTC GGA TCC TAA TAC GAC TCA CTA TAG GAG GAT GTA AAC TTA CAT A-3’, orf1b reverse primer, 5’ -CTC GAA TTC ATA GCT GGA TCA GCA GCA-3’ . T7 RNA promoter sequences are underlined and restriction sites for EcoRI and BamHI are italicized. PCR products were digested with BamHI and EcoRI and sub-cloned into BamHI and EcoRI site of pUC19. pMIF-cGFP-Zeo-hsa-miR-10b was purchased from System Biosciences (Mountain View, CA) and pSIF-Neo-lres-GFP-has-miR-21 was a gift from Dr. Yong Li (University of Louisville, USA). These were used as templates for PCR amplification to generate DNA templates suitable for direct use in in-vitro transcription. The PCR primers used were: pre-miR-10b forward primer, 5’ -GGA TCC TAA TAC GAC TCA CTA TAG GTA CCC TGT AGA ACC GAA T-3’, pre-miR-10b reverse primer, 5’ -ATT CCC CTA GAA TCG AAT-3’ ; pre-miR-21 forward primer, 5’ -GGA TCC TAA TAC GAC TCA CTA TAG GTA GCT TAT CAG ACT GAT G-3’, pre-miR-21 reverse primer, 5’ -ACA GCC CAT CGA CTG GTG-3’ . The plasmid pCYPAC2-CD44 which contains the last CD44 exon was a gift from Dr. Finn C. Nielsen (University of Copenhagen, Denmark), and was used for PCR amplification. The PCR primers used to amplify segment (nts 2861-3055) corresponding to 3’ untranslated region of CD44 RNA were: CD44 forward primer, 5’ -GGA TCC TAA TAC GAC TCA CTA TA GG AAA TTA GGG CCC MT TAA-3’, CD44 reverse primer, 5’ -AAA TTT CCT CCC AGG GAG-3’.
Preparation of radiolabeled nucleic acids
To synthesize spike, orf3, and orf1b RNAs, pUC19-Spike, pUC19-orf3, and pUC19-orf1b were each linearized with EcoRI and in-vitro transcribed using T7 RNA polymerase using previously described protocols [10]. To synthesize 5’ GG-pri-hsa-miR-10b, 5’ GG-pri-hsa-miR-21, 5’ GG-pre-hsa-miR-10b, 5’ GG-pre-hsa-miR-21 and CD44 RNA (nts 2862-3055), the PCR amplified DNA templates were used directly for in-vitro transcription by T7 RNA polymerase. All RNAs were then 5’ -radiolabeled with Y-[32P]-ATP using T4 polynucleotide kinase as previously described [17].
In vitro assay for endonuclease activity
The standard 20 μl-reaction mixture used for this assay included 2 mM DTT, 1.0 unit of RNa-sin, 2 mM magnesium acetate, 10 mM Tris-HCI, pH 7.4, and 1 ng of 5’ -32P -radiolabeled RNA (~5 × 104 cpm). The pH of all buffers for experiments described here was determined at room temperature. Reactions were incubated for 25 min at 37°C unless otherwise indicated. Forty μl of loading dye (9 M urea, 0.2% xylene cyanol, 0.2% bromophenol blue) were added to the reaction samples, and then 10 μl of reaction mixture were subjected to electrophoresis in 8% polyacrylamide, 7 M urea gel. For identification of cleavage sites, partial RNase T1 digestion and alkaline hydrolysis of radiolabeled RNA were done as previously described and samples were electrophoresed on 12% polyacrylamide/7 M urea gel [17]. Gels were then dried and subjected to phosphorlmaging using a Cyclone Phosphorlmager.
In vitro assay with APE1 and DICER
The reaction was carried out in two steps. First, a 10 μl-reaction mix containing 1 μM of recombinant APE1 and 25 nM of 5'-32P-labeled in vitro transcribed pre-miRNA analog (50,000 cpm/μl) in 8.8 mM Tris-HCI pH 7.4 and 2 mM magnesium acetate was incubated at 37°C for 30 min. Recombinant APE1 was purified and prepared in 20 mM Tris-HCI pH 8, 300 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 50% glycerol was used in this reaction. Second, 10 μl mixture containing 0.4 x DICER Reaction Buffer, 0.25 U DICER (Applied Biosystems, Ontario), and DEPC treated water was added to the first reaction and incubated at 37°C for 30 min. Addition of 0.4 x DICER Reaction Buffer (120 mM NaCl, 8 mM HEPES, 2 mM MgCl2, and 20 mM Tris-HCl pH 9.0) gives the final reaction mixture with salt and pH condition similar to that of the 1 x DICER Reaction Buffer which allows for optimal DICER activity. DICER was prepared to 0.25U/μl from 1 U/μl stock in storage buffer (100 mM NaCl, 20 mM Tris (pH7.5), 1 mM EDTA, and 50% glycerol) before combining with the DICER Reaction Buffer. The 0.4 x DICER Reaction Buffer was prepared from 5 X DICER Reaction Buffer (1.5 M NaCl, 100 mM HEPES, 25 mM MgCl2, and 250 mM Tris-HCl pH 9.0). The reaction was stopped by adding 40 μl of the Stopping Dye (9 M urea, 0.01% bromophenol blue, 0.01% xylene cyanol FF) and 6 μl was loaded onto the 8% polyacrylamide gel.
Secondary structure determination using RNase probes
A partial alkaline digestion of the 5'-radio-labeled transcript was carried out to generate an RNA ladder used to identify the cleavage sites. A typical reaction mixture of 20 μl included 5'-radiolabeled transcript (100,000 cpm), 50 mM sodium bicarbonate pH 9.2, and 1 mM EDTA. The reaction was carried out at 95°C for 3 min followed by incubation on ice for 1 min. RNase T1 (GE Healthcare), RNase A (Sigma), RNase VI (Ambion) and RNase T2 (Invitrogen) were used for enzymatic structure probing. Prior to probing, 5’ -32P-radiolabeled RNA (30,000 cpm) was taken through a refolding procedure which heated the RNA at 50-55° C for 5 min and then cooled to room temperature for 10 min. Digestion by RNase T1 was done under both native and denaturing conditions in order to determine if a particular guanosine residue is in single-stranded or stem region. Digestion of the 5’ -32P-radiolabeled transcript under denaturing condition was done in a 20 μl of 1 x sequencing buffer (Ambion, Inc., Austin, Texas) containing 20 mM sodium citrate pH 5, 7 M urea, and 1 mM EDTA. RNase T1 has been reported to be active under such condition [18]. Digestion of the transcript under native condition was done in a buffer containing 100 mM Tris-HCI pH 7.5, 1 M NH4Cl, and 100 mM MgCl2. One unit of RNase T1 was added and then incubated for 5 min at room temperature, while for partial digestion under RNA native conditions, 0.05 units of RNase T1 was added and incubated for 30 seconds on ice. Digestions with RNase T2 (0.02 units) and RNase A (2 x 10-4 mg) were performed for 15 min at room temperature, whereas for RNase V1 (1 x 10-4 units), incubation was done for 5 min at room temperature. Digestions with these RNases were carried out in 1 x Structure Buffer (Ambion, Inc., Austin, Texas) containing 10 mM Tris pH 7, 100 mM KCl, and 10 mM MgCl2. All reactions were terminated by the addition of 40 μl of loading dye (9 M urea, 0.2% xylene cyanol, 0.2% bromophenol blue). To check for RNA integrity, undigested RNA was also run concurrently (input RNA). Ten μl of probing reactions were visualized on 12% polyacrylamide/7M urea gels which were electrophoresed at a constant current (45 mA). Gels were dried and subjected to phosphorlmaging.
Results
Divalent metal ion requirements of APE1
We have previously demonstrated that the specificity of the endoribonuclease activity exhibited by the purified recombinant wild-type APE1 and absence of the enzymatic activity amongst other recombinant APE1 mutants, such as H309N and E96A, which were purified using an identical procedure [10]. In this study, we perform further biochemical characterization of the purified recombinant wild-type APE1.
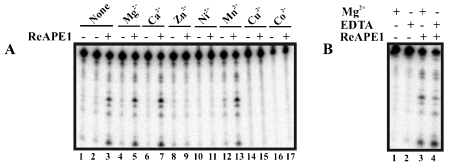
The AP DNA endonuclease activity of APE1 is divalent metal ion dependent [19–22]. We therefore determined the divalent metal ion requirement for RNA cleavage by APE1. For the purpose of the following experiments, protein samples were dialyzed in a metal ion-free buffer, as described in Materials and Methods, to ensure complete removal of metal ions. Figure 1A shows that APE1 was able to cleave c-myc CRD RNA in the absence of any divalent metal ions (lanes 1-3). RNA-cleaving activity was still visible when 10 mM EDTA was added (Figure 1B) further confirming APE1's divalent metal ion-independency. APE1 also exhibited activity in the presence of 2 mM Mg2+ (lanes 4-5), Ca2+ (lanes 6-7), and Mn2+ (lanes 12-13). In contrast, at 2 mM, Zn2+ (lanes 8-9), Ni2+ (lanes 1011), Cu2+ (lanes 14-15), and Co2+ (lanes 16-17), all had inhibitory effect on RNA-cleaving activity of APE1.
Figure 1.
Effect of divalent metal ions on the RNA-cleaving activity of APE1. (A) 350 fmoles of 5’ -32P-radiolabeled c-myc CRD RNA were treated with 1.4 μM of purified APE1 for 5 min at 37 °C in the presence (lanes 4-17) or absence (lanes 1-3) of 2 mM divalent metal ions as indicated under the in vitro endonuclease assay condition described in the Materials and Methods. Samples were electrophoresed on 8% polyacrylamide/7 M urea gel. (B) Experiment was performed as in (A) with the exception that 10 mM EDTA was present (lanes 2 and 4) or absent (lanes 1 and 3) in samples. 2 mM Mg2+ was included in samples in lanes 1 and 3.
APE1 cleaves RNA components of SARS-corona virus at specific sites
The purified recombinant human APE1 has thus far been tested only against c-myc CRD RNA [10]. To determine if APE1 can also cleave other RNAs, to further confirm the sequence and structural cleavage specificity of APE1, and to initiate studies into investigating the possible use of the enzyme as an anti-viral agent, we challenged APE1 with three RNA components, spike, orf1b and orf3 RNAs, which code for proteins important in the life cycle of the SARS-corona virus [23]. Spike or S protein mediates membrane fusion and binding of the virus to host receptors [23]. Orf1b is one of the replicases essential for replication of the virus [23]. Orf3, expressed during SARS-CoV infection, is a protein with endocytotic properties and does not have homology to other known coronaviruses [23].
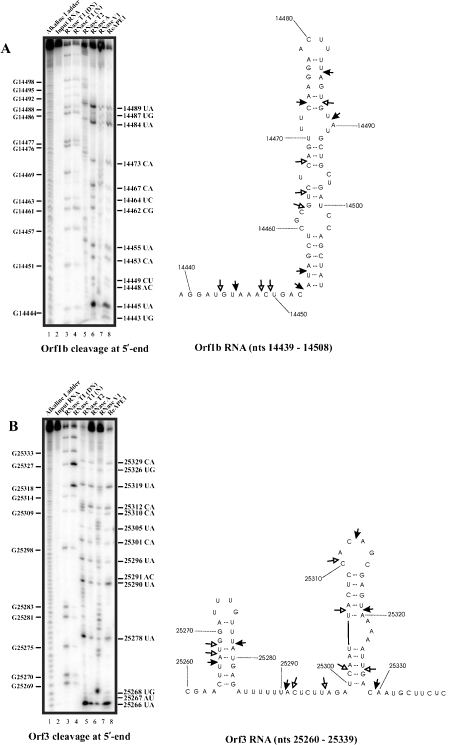
The secondary structure of orf1b RNA nts 14439-14508 was predicted using the software M-fold program [24], and the RNase structure probing data using RNases T1, T2, A, and V1, as shown in the left panel of Figure 2A were then used as a constraint with the structure generated by Mfold to obtain a structure that best fit the probing data (right panel, Figure 2A). APE1 cleavage sites on orf1b RNA were then mapped as shown in Figure 2A (lane 8 in the left panel). Moderate to major cleavage sites generated by APE1 are 14445UA, 14453CA, 14455UA, 14473CA, 14484UA, and 14489UA, while weak cleavages are at 14443UG, 14448AC, 14449CU, 14462CG, 14464UC, 14467CA, and 14487UG. Several key observations can be drawn from this result: (i) there is a preference for UA over UG as exemplified by more intense cleavage at 14445UA over 14443UG, (ii) cleavages can occur at CA, UA, and UC at weakly paired regions as exemplified by 14464UC, 14467CA, 14473CA, and 14484UA, (iii) for the first time, APE1 is shown to cut very weakly at CU, AC and CG at single-stranded region as exemplified by 14448AC, 14449CU, and 14462CG sites.
Figure 2.
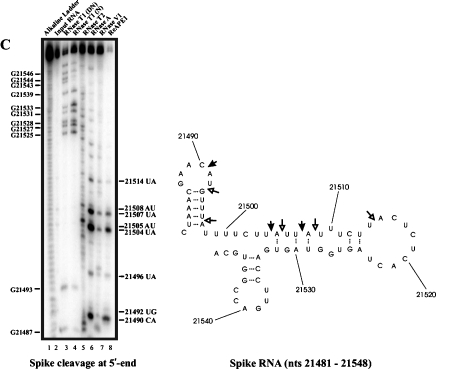
APE1 cleaves spike, orf3, and orf1b RNA components of SARS-Corona virus at specific sites. Ribonuclease secondary structure probing of orf1b (A), orf3 (B), and spike (C) RNA. In vitro transcribed RNA corresponding to the indicated nucleotides was 5’ -end-labelled with 32P and refolded prior to probing with the indicated RNases. For reference, an alkaline hydrolysis ladder was generated as shown (lane 1). Numbering on the left indicates guanosine residue sites cleaved by RNase T1 under denaturing conditions, whereas numbering on the right indicates sites cleaved by APE1. The RNA secondary structure model on the right panel shows locations of sites cleaved by APE1. The structure was obtained by using the probing data on the left panel as constraints in the Mfold program [24]. Filled arrows represent strong cleavage while un-filled arrows represent weak cleavage. DN denotes cleavage by RNase T1 under denaturing conditions, and N denotes cleavage by RNase T1 under native conditions.
Similarly, the secondary structure of orf3 RNA nts 25260-25339 was predicted using M-fold program [24], and the RNase structure probing data as shown in the left panel of Figure 2B were then used to generate a structure that best fit the probing data (right panel, Figure 2B). Moderate to strong cleavage by APE1 are at the following sites: 25266UA, 25278UA, 25290UA, 25312CA, 25319UA, and 25329CA. Weaker but significant cleavage sites are observed at 25267AU, 25268UG, 25291AC, 25296UA, 25301CA, 25305UA, 25310CA, and 25326UG. The following observations are noted: (i) cleavage at UA sites at weakly paired regions as exemplified by 25266UA, 25278UA, and 25319UA, (ii) APE1 cuts weakly at AC as exemplified by a cleavage at 25291AC, (iii) APE1 can cut weakly at stem region as exemplified with 25267AU, 25268UG, and 25301CA.
The secondary structure of spike RNA nts 21481-21548 was also predicted using M-fold program [24], and the RNase structure probing data were then used as a constraint with the structure generated by Mfold to obtain a structure that best fit the probing data (right panel, Figure 2C). In the 68 nt spike RNA, APE1 cleaves strongly at 21490CA, 21504UA, and 21507UA while weaker cleavages are seen at 21492UG, 21496UA, 21505AU, 21508AU, and 21514UA. The following are key observations from this result: (i) APE1 can cut weakly at UA at stem region as exemplified by a weak cleavage at 21496UA, (ii) APE1 can cut weakly at AU sites at weakly paired region as exemplified by cleavages at 21505AU and 21508AU.
APE1 cleaves 3’ UTR of CD44 RNA at specific sites
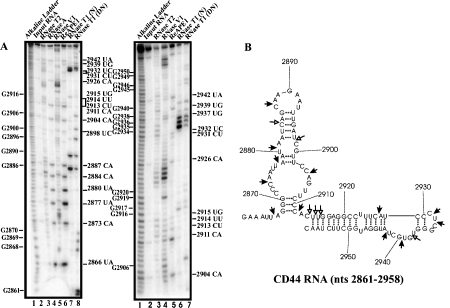
Multiple sites at 3’ UTR of CD44 mRNA have been shown to bind proteins and following depletion of the RNA-binding proteins, CD44 mRNA became unstable [25]. Thus, it was postulated that the RNA-binding proteins bind to and protect CD44 mRNA from endonucleolytic cleavage. To determine if APE1 can cleave a segment of 3’ UTR of CD44 mRNA that binds RNA-binding protein and to further confirm the sequence and structural cleavage specificity of APE1, we challenged in vitro transcribed CD44 RNA nts 2861-3055 with the purified recombinant APE1. The secondary structure of CD44 3’ UTR RNA nts 2861-3055 was first predicted using Mfold program [24]. RNase probing experiments were then conducted (left panel, Figure 3) and the results were used as a constraint with the structure generated by Mfold to obtain a structure that best fit the probing data. Only the RNA structure encompassing nts 2861-2958 is shown (right panel, Figure 3). As shown, the major cleavage sites generated by APE1 (lane 6 on the left panel gel; lane 5 on the right panel gel) are 2866UA, 2873CA, 2877UA, 2880UA, 2884CA, 2887CA, 2904CA, 2911CA, 2926CA, 2931CU, 2932UC, 2939UG, and 2942UA. Weak but distinct cleavage sites generated by APE1 are 2898UC, 2913CU, 2914UU, 2915UG, and 2937UG. The following are key observations from this experiment: (i) APE1 does cut CA at stem regions as exemplified by cleavages at 2884CA and 2926CA, (ii) cleaves at UG at weakly paired regions as shown by cleavage at 2915UG, (iii) cleaves at CU and UC at a single-stranded region as exemplified by cleavage at 2931CU and 2932UC.
Figure 3.
APE1 cleaves 3’ UTR of CD44 RNA at specific sites. Ribonuclease secondary structure probing of CD44 RNA was done as described in Fig. 2. Numbering on the left indicates guanosine residue sites cleaved by RNase T1 under denaturing conditions, whereas numbering on the right indicates some of the sites cleaved by APE1. The RNA secondary structure model on the right panel shows the locations of sites cleaved by APE1. Filled arrows represent strong cleavage while un-filled arrows represent weak cleavage. DN denotes cleavage by RNase T1 under denaturing conditions, and N denotes cleavage by RNase T1 under native conditions.
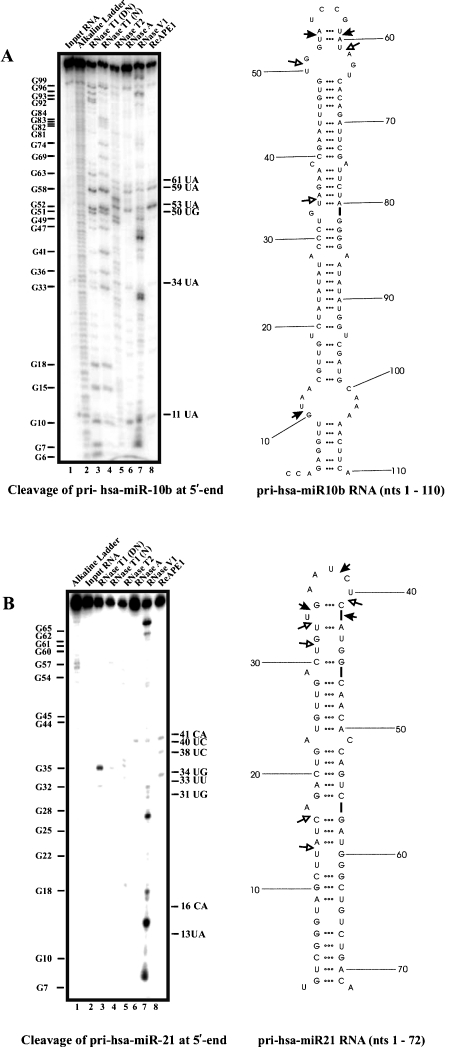
APE1 cleaves pri-miR-21 and pri-miR-10b at specific sites
In our effort to establish the RNA sequence and structural cleavage specificity of APE1 and to initiate studies into the possible use of APE1 in degrading oncogenic miRNAs, we challenged in vitro transcribed pri-miR-21 and pri-miR-10b with purified recombinant human APE1. The secondary structure of pri-miR-10b was predicted using Mfold program [24], and as before, RNase probing experiments were conducted. The data was used as a constraint with the structure generated by Mfold to obtain a structure that best fit the probing data (right panel, Figure 4A). The major cleavage sites generated by APE1 are 11UA, 53UA, and 59UA, while weaker sites are observed at 34UA, 50UG, and 61UA. Key observations from this result are: (i) APE1 cleaves UA at weakly paired regions as shown by cleavages at 34UA and 53UA, (ii) APE1 does not cut at UA, CA, UG, and UC at strong stem regions as exemplified by the absence of cleavage at 21UA, 23UA, 25UA, 48UG, 65CA, 67CA, and 71UC.
Figure 4.
APE1 cleaves 5’ GG-pri-hsa-miR-10b and 5’ GG-pri-hsa-miR-21 at specific sites. Ribonuclease secondary structure probing of 5’ GG-pri-hsa-miR-10b (A) and 5’ GG-pri-hsa-miR-21 (B) was done as described in Fig. 2. Numbering on the left indicates guanosine residue sites cleaved by RNase T1 under denaturing conditions, whereas numbering on the right indicates some of the sites cleaved by APE1. The RNA secondary structure model on the right panel shows the locations of sites cleaved by APE1. Filled arrows represent strong cleavage while un-filled arrows represent weak cleavage. DN denotes cleavage by RNase T1 under denaturing conditions, and N denotes cleavage by RNase T1 under native conditions.
Similarly, RNase probing experiments were performed on pri-miR-21 and the secondary structure generated upon constraining the experimental data with structure predicted by Mfold, is shown on the right panel of Figure 4B. As shown, the major cleavage sites generated by APE1 are 34UG, 38UC, and 41CA, while weaker cleavage sites are seen at 13UA, 16CA, 31UG, 33UU, and 40UC. The following are key observations from this result: (i) APE1 does not cut at UG, CA, and UU at strong stems as indicated by the absence of cleavage at 12UU, 24UG, 26UU, 27UG, 43UG, 46CA, 49CA, 52CA, and 55UC.
Pre-treatment with APE1 interferes with the processing of pre-miR-10b and pre-miR-21 by Dicer
To assess if APE1 can potentially interfere with the processing of pre-miRNAs by Dicer, we treated pre-miR-10b and pre-miR-21 with APE1 prior to the incubation with Dicer. It is important to point out that two additional nucleotides GG was inserted 5’ to pre-miR-10b and pre-miR-21 DNA templates which is necessary for T7 in vitro transcription. However, the resulting RNA substrates, 5'GG-pre-hsa-miR-21 (Figure 5) and 5'GG-pre-has-miR-10b (Figure 6), have identical secondary structure as their in vivo pre-miRNA substrates as predicted by Mfold. Hence, APE1 cleavage sites are expected to be identical on both types of substrates.
Figure 5.
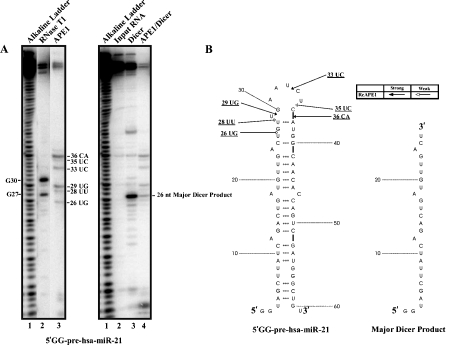
Effect of pre-treatment with APE1 on Dicer's ability to process 5’ GG-pre-hsa-miR-21. (A) 350 fmoles of 5’ -32P-radiolabeled 5’ GG-pre-hsa-miR-21 were treated with 1.4 μM of purified APE1 (lane 3, left panel gel) for 5 min at 37°C under the in vitro endonuclease assay condition as described in the Materials and Methods. For reference, an alkaline hydrolysis ladder was generated (lane 1, left and right panel gels). For the left panel gel, numbering on the left indicates guanosine residue sites cleaved by RNase T1 under denaturing conditions (lane 2), whereas numbering on the right indicates sites cleaved by APE1. For the right panel gel, the numbering on the right indicates the major cleavage product generated by Dicer. 5’ -32P-radiolabeled 5’ GG-pre-hsa-miR-21 pre-treated with (lane 4, right panel gel) or without (lane 3, right panel gel) APE1 prior to the incubation with 0.25 units of Dicer for 30 min at 37°C. (B) The RNA secondary structure of 5’ GG-pre-hsa-miR-21 shows the locations of cleavage sites generated by APE1. The major Dicer product is shown.
Figure 6.
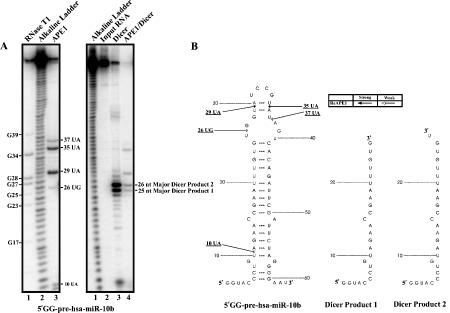
Effect of pre-treatment with APE1 on Dicer's ability to process 5’ GG-pre-hsa-miR-10b. (A) 350 fmoles of 5’ -32P-radiolabeled 5’ GG-pre-hsa-miR-10b were treated with purified APE1 (lane 3, left panel gel) for 5 min at 37°C under the in vitro endonuclease assay condition as described in the Materials and Methods. For reference, an alkaline hydrolysis ladder was generated (lane 2, left panel gel; lane 1, right panel gel). For the left panel gel, numbering on the left indicates guanosine residue sites cleaved by RNase T1 under denaturing conditions (lane 1), whereas numbering on the right indicates sites cleaved by APE1. For the right panel gel, the major cleavage products 1 and 2 generated by Dicer are shown with numbering on the right. 5’ -32P-radiolabeled 5’ GG-pre-hsa-miR-10b pre-treated with (lane 4, right panel gel) or without (lane 3, right panel gel) APE1 prior to incubation with 0.25 units of Dicer for 30 min at 37°C. (B) The RNA secondary structure of 5’ GG-pre-hsa-miR-10b shows the locations of cleavage sites generated by APE1. Dicer products 1 and 2 are shown.
The left panel in Figure 5 (lane 3 on left panel gel) shows APE1 cleaves pre-miR-21 at 26UG, 28UU, 29UG, 33UC, 35UC, and 36CA. Dicer cleaves pre-miR-21 to generate a single major product of 26 nt in size as shown in Figure 5 (lane 3 on right panel gel). Pre-treatment with APE1 significantly reduce the ability of Dicer to process pre-miR-21 as evident by the significant reduction (~6.2 fold) of the 26 nt major Dicer product. The left panel in Figure 6 shows the cleavage of APE1 on pre-miR10b at 26UG, 29UA, 35UA, and 37UA. On the other hand, Dicer alone cleaves pre-miR-10b to generate two major products, 25 nt Product 1 and 26 nt Product 2 (lane 3 on the right panel gel). Pre-treatment of pre-miR-10b with APE1 appeared to substantially suppress the ability of Dicer to process pre-miR-10b, as evident from the significant reduction in Products 1 (~6.2 fold) and 2 (~6.7 fold) (lane 4 on the right panel gel).
Discussion
Given that APE1 could act as an important endoribonuclease to control gene expression in cells, it is thus warranted to further understand the RNA-cleaving properties of APE1. Prior to this study, several RNA-cleaving properties of APE1 were unknown. First, it was unknown whether APE1 required the presence of divalent metal ions for its RNA catalysis. Second, APE1 was found to preferentially cleave c-myc CRD RNA at UA, UG, and CA dinucleotides [10]. However, it was unknown whether APE1 can cleave these or other sequences on other RNA substrates that have biological significance.
Many nucleases that degrade DNA or RNA utilize metal ions for their catalysis [26]. Against a standard abasic DNA substrate, APE1 requires divalent metal ions for its optimal activity [20,27]. It is reported that these metal ions occupy the active site to stabilize the transitional state during DNA incision, and facilitate the release of the incised product. Previously, it was found that Mg2+, Mn2+, Ni2+, and Zn2+ was able to rescue an EDTA-inactivated APE1 incision activity on an abasic DNA substrate while Ca2+ could not [3]. Another study found Ni2+ and Zn2+ ions permitted the DNA incision activity up to concentrations of 100 μM (Ni2+) and 50 μM (Zn2+) [32]. Comparing these requirements with our data on the ability of APE1 to cleave c-myc CRD RNA, we found a few notable discrepancies. First, we found that APE1 was able to cleave RNA without the presence of divalent metal ions. Possible explanation may be that APE1 has previously been found to weakly cleave abasic DNA in the presence of EDTA [21], and in particular, it has been shown to have an EDTA-resistant activity towards acyclic substrates that are considered to have a better flexibility than abasic DNA [20]. Single stranded RNA structures (e.g., hairpin loops) are particularly more flexible than those of RNA stems and abasic DNAs, which could allow APE1 to cleave such RNA substrates without their divalent metal cofactors. Secondly, we showed that APE1 is able to cleave RNA in the presence of Ca2+. Here, the discrepancy between the RNA and DNA cleaving may be that Ca2+ is not suitable to occupy the APE1 active site for DNA incision because of its larger atomic size than Mg2+, but for RNA cleaving, such metal ions are not required. Thirdly, we showed the RNA-cleaving activity of APE1 is inhibited by the divalent metal ions Ni2+and Zn2+, which are known to interact with histidines to inhibit the enzymatic activities of certain proteins [29,30]. The presence of these ions may thus affect the amino acid residues that are specifically critical for RNA catalysis of APE1. Alternatively, these metal ions may affect the secondary structure of RNA substrates and interfere with APE1 binding to these RNAs or interfere with their release after incision. Further experimentations are clearly required to answer these questions.
We found that APE1 is able to cleave RNAs in a non-specific manner in vitro. This includes miRNAs, CD44 RNA, and RNA components for SARS-corona virus. We propose that in addition to mRNA, APE1 may have multiple RNA substrates in vivo. Similar to our previous finding [10], APE1 cleavage sites were found to be predominantly located in the single-stranded regions of the RNA substrate with dinucleotide sequences of UA, UG, and CA. In addition, for the first time, we found that APE1 can weakly cleave RNA at UC, CU, AC, and AU sites. APE1 has previously been shown to cleave abasic RNA [31,32] and is involved in ribosomal RNA quality control process [32]. Although the N-terminus of APE1 appears to be important in binding RNA and in abasic RNA-cleaving function [32], the detailed structural basis for this and the endoribonuclease activity of APE1 reported here remain unclear. In abasic DNA binding, APE1 interacts strongly with an abasic deoxyribose flipped out of the DNA helix and it tightly binds to this moiety facilitated by a hydrophobic pocket composed of Phe 266, Trp 280,and Leu 282. Such tight packing expels the binding of regular nucleotides and gives specificity towards the abasic sites [33]. However,since all RNA molecules tested possess bases, the likelihood of such specificity is low and it is likely that APE1-RNA interaction is more tolerant towards different nucleotides than its interaction with an abasic DNA. This would result in range of specificities cleaving different dinucleotides sequences. Single stranded regions of RNA, as in the case of RNA hairpin loops, have their bases flipping outwards from the centre of the loop which allow RNA binding proteins to interact easily [34]. In RNA cleaving, we speculate that APE1 can recognize some of these extra helical bases in single stranded RNAs. The APE1 residues which confer specificity for the 3’ of pyrimidines still need to be identified. But, some insights can be drawn from the example of RNase A. In recognition of its substrate, RNase A uses residue Thr 45 for recognition to cleave polyC and polyU [35]. Such specificity is facilitated by specific hydrogen bonds between the side chain -OH with -NH (pyrimidine) and the main chain -NH and =O (pyrimidine). In APE1 RNA cleavage specificity, Thr 268 could serve a similar purpose, which is located near the hydrophobic pocket of APE1 (Phe 266-Trp 280-Leu 282) for recognizing the extra helical abasic deoxyribose [33]. Site-directed mutagenesis studies are clearly required to test this hypothesis. After testing different RNA substrates with varying secondary structures, we conclude that APE1 cleaves only on the single stranded regions or weakly paired regions of the RNA. It has strong cleavage specificity towards 3’ of pyrimidines, especially at UA, UG, and CA sites.
Since CA-rich elements are known as potent splicing enhancers/silencers [36], the preference of APE1 to cleave CA dinucleotides suggests a possible role for APE1 in mRNA splicing. This is in line with the finding that APE1 serves as a nuclear enzyme and has been found to interact with heterogenous nuclear ribonucleo-protein L (hnRNP L) [37], a high affinity CA-rich/CA-repeat binding protein which acts as a key regulator of splicing [38–40]. In addition, APE1 was found to interact with YB-1 [41], a mediator of splicing [42].
Pre-treatment of pre-miRNAs with APE1 significantly reduces the ability of Dicer to process these substrates in vitro. This provides another dimension in which APE1 could potentially influence the biogenesis and hence the level of miRNAs in cells. Studies are currently underway to test this hypothesis. In summary, we have uncovered some important biochemical properties of APE1. APE1 cleaves RNA without the presence of divalent metal ions. APE1 is able to cleave any RNA when presented in vitro and it only cleaves in the single-stranded regions or weakly paired regions. It prefers to cleave 3’ of pyrimidines at UA, UG, and CA sites. In addition, although weakly, APE1 can cleave at UC, CU, AC, and AU sites.
Acknowledgements
We thank Dr. Wai Ming Li for critical comments on this manuscript and Dr. Sankar Mitra for the plasmid pET15b-APE1.
This work was supported by a grant from Natural Science & Engineering Research Council (Grant # 227158) to C.H.L. W.C.K. was a recipient of Pacific Century Scholarship, Michael Smith Foundation of Health Research Junior Graduate Scholarship Award, and NSERC Canada Graduate Scholarship. D.K. was a recipient of NSERC Undergraduate Student Research Awards.
Abbreviations
- APE1
Apurinic/apyrimidinic endonuclease 1
- CRD
coding region determinant
- 3'UTR
3'-untranslated region
- RNase
ribonuclease
References
- [1].Wilson DMIII, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- [2].Chen DS, Herman T, Demple B. Two distinct human DNA diesterases that hydrolyze 3(-bloking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Barzilay G, Walker U, Robson CN, Hickson ID. Site-directed mutagenesis of the human DNA repair enzyme HAP1: identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res. 1995;23:1544–1550. doi: 10.1093/nar/23.9.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kane CM, Linn S. Purification and characterization of an apurinic/apyridiminic endonuclease from HeLa cells. J Biol Chem. 1981;256:3405–3414. [PubMed] [Google Scholar]
- [5].Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- [6].Wilson DMIII, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- [7].Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–656. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–576. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- [9].Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of apurinic/apyrimidinic endonuclease APE1 as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Herrick DJ, Ross J. The half-life of c-myc mRNA in growing and serum-stimulated cells: influence of the coding and 3’ untranslated regions and role of ribosome translocation. Mol Cell Biol. 1994;14:2119–2128. doi: 10.1128/mcb.14.3.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yielding NM, Lee WMF. Coding elements in exons 2 and 3 target c-myc mRNA downregulation during myogenic differentiation. Mol Cell Biol. 1997;17:2698–2707. doi: 10.1128/mcb.17.5.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bernstein PL, Herrick DJ, Prokipcak RD, Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region determinant. Genes Dev. 1992;6:642–654. doi: 10.1101/gad.6.4.642. [DOI] [PubMed] [Google Scholar]
- [14].Morello D, Lavenu A, Pournin S, Babinet C. The 5’ and 3’ non-coding sequences of the c-myc gene, required in vitro for its post-transcriptional regulation, are indispensable in vivo. Oncogene. 1991;8:1921–1929. [PubMed] [Google Scholar]
- [15].Sambrook J, Russell DW. CSHL Press, Cold Spring Harbor: New York; 2001. Molecular Cloning - A Laboratory Manual (3rd ed.) [Google Scholar]
- [16].Marra M, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, Chan SY, Cloutier A, Coughlin SM, Freeman D, Girn N, Griffith OL, Leach SR, Mayo M, McDonald H, Montgomery SB, Pandoh PK, Petrescu AS, Robertson AG, Schein JE, Siddiqui A, Smailus DE, Stott JM, Yang GS, Plummer F, An-donov A, Artsob H, Bastien N, Bernard K, Booth TF, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples GA, Tyler S, Vogrig R, Ward D, Watson B, Brunham RC, Krajden M, Petric M, Skowronski DM, Upton C. Roper RL The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- [17].Bergstrom K, Urquhart JC, Tafech A, Doyle E, Lee CH. Purification and characterization of a novel mammalian endoribonuclease. J Cell Biochem. 2006;98:519–537. doi: 10.1002/jcb.20726. [DOI] [PubMed] [Google Scholar]
- [18].Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Beernink PT, Segelke BW, Hadi MZ, Erzberger JP, Wilson DMIII, Rupp B. Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, APE1: implications for the catalytic mechanism. J Mol Biol. 2001;307:1023–1034. doi: 10.1006/jmbi.2001.4529. [DOI] [PubMed] [Google Scholar]
- [20].Erzberger JP, Wilson DMIII. The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J Mol Biol. 1999;290:447–457. doi: 10.1006/jmbi.1999.2888. [DOI] [PubMed] [Google Scholar]
- [21].Masuda Y, Bennett RAO, Demple B. Rapid dissociation of human apurinic endonuclease (Ape 1) from incised DNA induced by magnesium. J Biol Chem. 1998;273:30360–30365. doi: 10.1074/jbc.273.46.30360. [DOI] [PubMed] [Google Scholar]
- [22].Oezguen N, Schein CH, Peddi SR, Power TD, Izumi T, Braun A. A ‘moving metal mechanism for substrate cleavage by the DNA repair endonuclease APE1. Proteins. 2007;68:313–323. doi: 10.1002/prot.21397. [DOI] [PubMed] [Google Scholar]
- [23].Tan YT, Lim SG, Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;15:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vikessa J, Hansen TVO, Jonson L, Borup R, Wewer UM, Christiansen J, Neilsen FC. RNA-binding IMPs promote cell adhesion and invado-podia formation. EMBOJ. 2006;25:1456–1468. doi: 10.1038/sj.emboj.7601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang W. An equivalent metal ion in one- and two -metal-ion catalysis. Nat Struct Mol Biol. 2008;15:1228–1231. doi: 10.1038/nsmb.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wilson DMIII. APE1 abasic endonuclease activity is regulated by magnesium and potassium concentrations and is robust on alternative DNA structures. J Mol Biol. 2005;345:1003–1014. doi: 10.1016/j.jmb.2004.11.028. [DOI] [PubMed] [Google Scholar]
- [28].Wang P, Guliaev AB, Hang B. Metal inhibition of human N-methylpurine-DNA glycosylase activity in base excision repair. Toxicol Lett. 2006;166:237–247. doi: 10.1016/j.toxlet.2006.06.647. [DOI] [PubMed] [Google Scholar]
- [29].Kang HW, Moon HJ, Joo SH, Lee JH. Histidine residues in the IS3-IS4 loop are critical for nickel -sensitive inhibition of the Cav2.3 calcium channel. FEBS Lett. 2007;581:5774–5780. doi: 10.1016/j.febslet.2007.11.045. [DOI] [PubMed] [Google Scholar]
- [30].Okamura M, Terada T, Katsura T, Saito H, Inui K. Inhibitory effect of zinc on PEPT1-mediated transport of glycylsarcosine and beta-lactam antibiotics in human intestinal cell line Caco-2. Pharm Res. 2003;20:1389–1393. doi: 10.1023/a:1025797808703. [DOI] [PubMed] [Google Scholar]
- [31].Berquist BR, McNeill DR, Wilson DMIII. Characterization of abasic endonuclease activity of human Ape 1 on alternative substrates, as well as effects of ATP and sequence context on AP incision. J Mol Biol. 2008;379:17–27. doi: 10.1016/j.jmb.2008.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vascotto C, Fantini D, Romanello M, Casaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D'Ambrosio CD, Scaloni A, Quadrifoglio F, Tell G. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mol CD, Izumi T, Mitra S, J.A. Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- [34].De Guzman RN, Turner RB, Summers MF. Pro-tein-RNA recognition. Biopolymers. 1998;48:181–195. doi: 10.1002/(SICI)1097-0282(1998)48:2<181::AID-BIP7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- [35].Raines RT, Ribonuclease A. Chem Rev. 1998;98:1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- [36].Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- [37].Kuninger DT, Izumi T, Papaconstantinou J, Mitra S. Human AP-endonuclease 1 and hnRNP-L interact with a nCaRE-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res. 2002;30:823–829. doi: 10.1093/nar/30.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hui J, Hung LH, Heiner M, Schreiner S, Neumüller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBOJ. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microar-ray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol. 2009;29:1442–1451. doi: 10.1128/MCB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, Kohno K, Mitra S, Bhakat KK. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28:7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Raffetseder U, Frye B, Rauen T, Jürchott K, Royer HD, Jansen PL, Mertens PR. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J Biol Chem. 2003;278:18241–18248. doi: 10.1074/jbc.M212518200. [DOI] [PubMed] [Google Scholar]