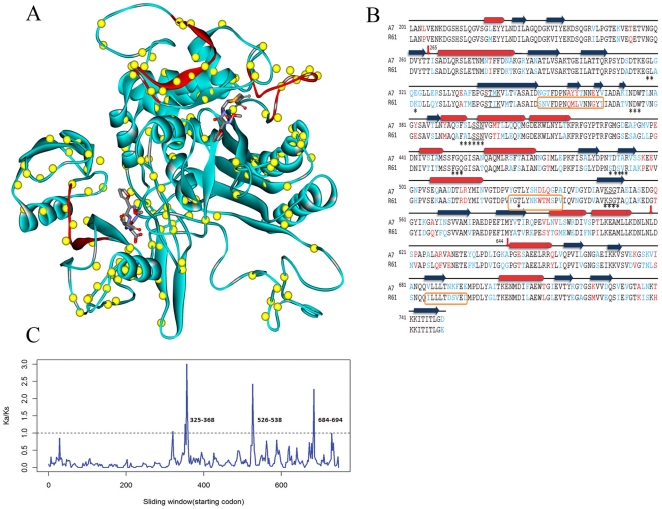
Figure 1. Comparison of PBP2x-A7 and PBP2x-R61.
(A). Structure of PBP2x from drug-resistant strain R61. Yellow spheres represent locations of mutations (as compared with the PBP2x sequence from the penicillin-sensitive strain A7). Two molecules of cefuroxime are placed in an equivalent position to that observed in the complex from PDB 1QMF. Three detected positive selection regions are colored in red. The result demonstrates that the three regions are all located in loops. Two of the three regions are adjacent to the upper cefuroxime molecule and surround the cleft of catalytic active site. The third positive selection region is far away from the active site and only located in the C-terminal domain. However, this region is very close to the lower cefuroxime molecule. (B). Sequence alignment of PBP2x-A7 and PBP2x-R61 and secondary structure assignment. Conserved mutations are represented in blue, and non-conserved changes are shown in red. Secondary structural elements in the PBP2x-R61 structure are shown as red cylinders (α-helices) and blue rectangles (β-sheets). Sequence motifs relevant for catalysis (SXXK, SXN and KSG) are underlined. The transpeptidase domain consists of residues 256–619. Residues that are less than 5 Å from the active site are marked with asterisks. Residues in regions that are under positive selection are shown within orange rectangular boxes. (C). Selection pressure on the PBP2x-R61 sequence. We used a sliding window of 10 codons (step size 2 codons) along the PBP2x-R61 DNA sequence to expose selective pressures on different regions. In the plot, although the general trend is below the threshold of 1 (indicated by a dotted line), there are 3 peaks over the threshold, indicating that these regions (residues 325–368,526–538 and 684–694) are under positive selection. Mutations in these regions appear to provide some fitness advantage to the bacterial strain.