Table 1.
Statistical evaluation of the structure calculation
| Final 25 structures | 2LD6a | |
|---|---|---|
| Rmsd from the distance constraints (Å) and dihedral angle constraints (°) | ||
Long-range NOE 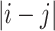 > 4 (462) > 4 (462) |
0.02708 ± 0.0045 | 0.020 |
Medium range NOE 1 ≤ 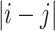 ≥ 4 (1,553) ≥ 4 (1,553) |
0.05088 ± 0.0010 | 0.048 |
| Intraresidue NOE (743) | 0.02256 ± 0.0030 | 0.016 |
| Hydrogen bond (140) | 0.01084 ± 0.0019 | 0.008 |
Dihedral angle 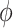 (85) [°] (85) [°] |
0.55 ± 0.09 | 0.541 |
| Rmsd form ideal geometry used in X-PLOR | ||
| Bond lengths (Å) | 0.0044 ± 0.0001 | 0.042 |
| Bond angle (°) | 0.68 ± 0.0077 | 0.682 |
| Improper angle (°) | 0.57 ± 0.0084 | 0.551 |
| Backbone helices | Side-chain | |
|---|---|---|
| Coordinate rmsd (Å) for non-hydrogen atoms in the final structures | ||
| Versus the averaged structure | 0.41 ± 0.033 | 0.94 ± 0.48 |
| Versus the averaged and minimized structure (2LD6) | 0.48 ± 0.28 | 1.08 ± 0.38 |
Numbers in parentheses indicate the total numbers of constraints in the categories
aThe average, minimized structure was derived from the final 25 structures. The thresholds of constraint violations reported here for NOE, dihedral angle and bond length were set at 0.5 Å, 5°, and 0.05 Å, respectively
