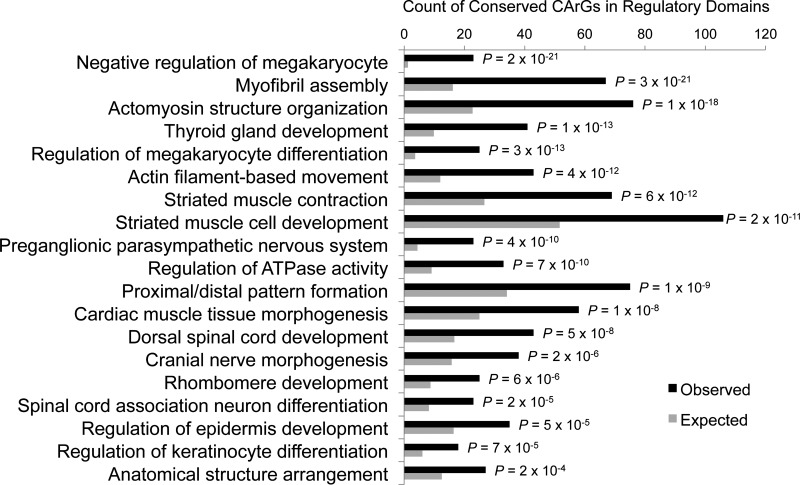
Fig. 4.
Genomic Regions Enrichment of Annotations Tool (GREAT) enrichment analysis of theoretical SRF-target genes based on biological process Gene Ontology (GO) terms. Enrichment analysis was performed using the GREAT algorithm (http://great.stanford.edu/) for each of the 8,252 conserved CArG sequences identified with the in silico method. The GREAT algorithm associates each human gene (n = 17,578) with a regulatory domain in the human genome (hg19 assembly) and calculates the total fraction of the genome annotated with GO terms (e.g., myofibril assembly). The submitted sequences that fall in each annotated GO term region are counted as “hits.” A binomial test compares the expected number of hits in a genome region with the observed number of hits. Listed in the figure are the most significantly enriched biological process GO terms (out of 7,170) for the conserved CArG sequences. Expected and observed counts for each GO term are listed with binomial test P value.