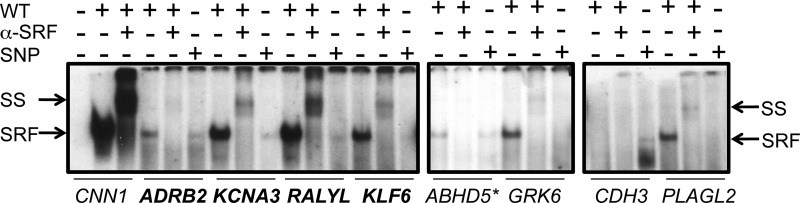
Fig. 6.
Altered SRF binding with SNPs in or near CArG boxes. In vitro translated (IVT) SRF was incubated with 32P-labeled wild-type (WT) or SNP mutant CArG boxes in close proximity to 8 putative SRF-dependent genes. SRF nucleoprotein complexes are indicated by the lower arrow. Verification of SRF binding was demonstrated by SRF antibody supershifting (SS) of the nucleoprotein complex (upper arrow). The known SRF target gene CNN1 was used as a positive control for SRF binding; human CNN1 does not have any known SNPs associated with its CArG box. Boldfaced gene names indicate CArGs that conform to the consensus sequence, while lightfaced gene names are CArG-like sequences that deviate from the consensus sequence by 1 bp. Seven of the putative SRF-target genes studied have the SNP located within their associated 10-bp CArG box. The final gene, ABHD5, has an SNP located 2 bp outside its' CArG box (highlighted with an asterisk). Data were replicated in an independent experiment. Exposure times were 24, 48, and 17 h, for each panel from left to right.