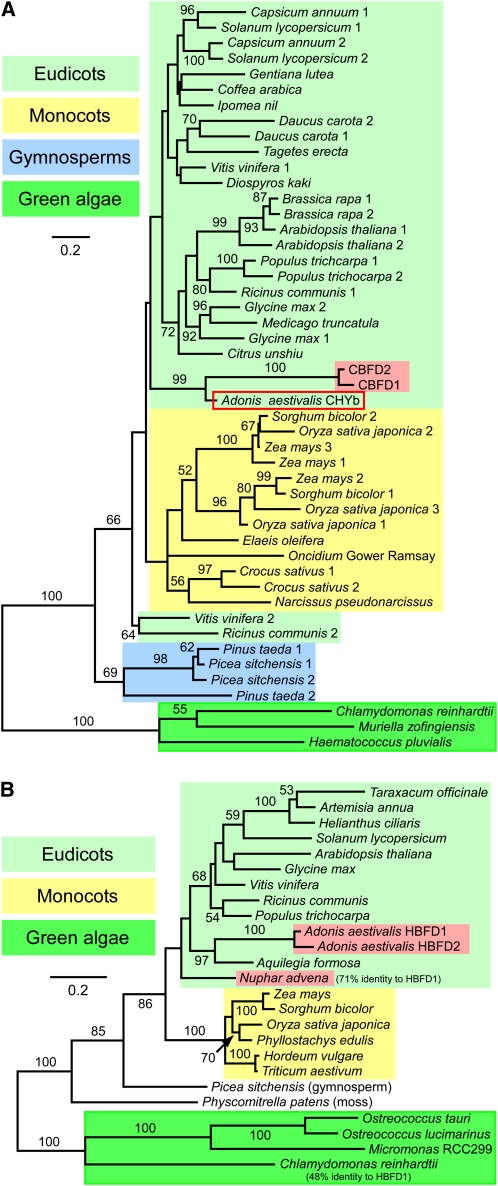
Figure 8.
Maximum Likelihood Trees for the CBFD and HBFD Enzymes of A. aestivalis and Related Polypeptides Encoded by cDNAs and Genes of Other Plants and Green Algae.
(A) Tree for plant and green algal carotenoid β-ring 3-hydroxylase enzymes of the membrane-integral, diiron, nonheme oxygenase type (CHYb-type) together with the A. aestivalis CBFD1 and CBFD2 polypeptides. CBFD1 and CBFD2 are highlighted with a red background. The A. aestivalis CHYb is enclosed in a red box. The amino acid sequences of CBFD1 and CBFD2 are ~90% identical to each other and ~58 to 60% identical overall to the A. aestivalis CHYb.
(B) Tree for the A. aestivalis HBFD1 and HBFD2 enzymes together with related polypeptides encoded by cDNAs and genes of other plants and green algae. HBFD1 and HBFD2 are highlighted with a red background, as is a N. advena polypeptide also shown to have 4-hydroxy-β-ring 4-dehydrogenase activity (Figure 9). Green algal sequences served as an outgroup for each tree. One thousand bootstrap trials were conducted, with bootstrap values >50% indicated. Alignments used to construct these trees are shown in Supplemental Figures 3 (for the CBFD and CHYb enzymes) and 4 (for HBFD and related polypeptides) online and are available in FASTA format as Supplemental Data Sets 1 and 2 online, respectively.