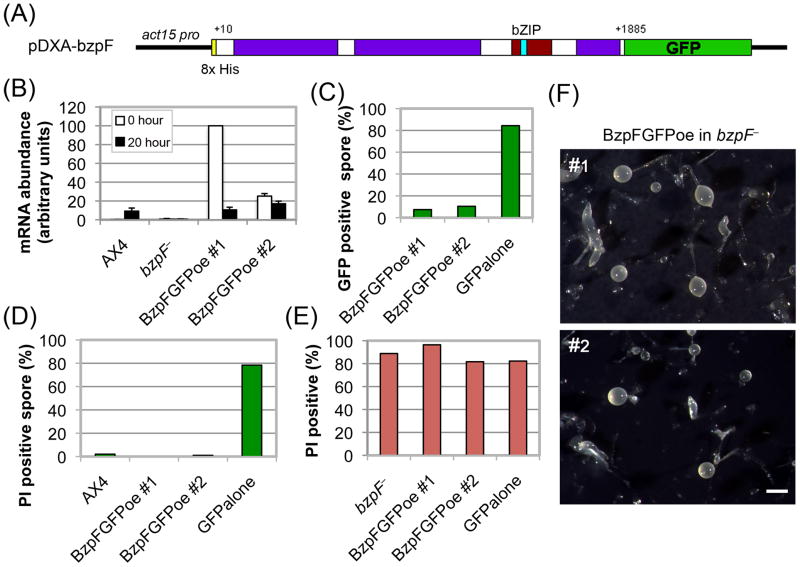
Fig. 5. Ectopic expression of bzpF complements the compromised spore phenotype.
(A) The BzpF ectopic expression construct: We fused most of the bzpF coding region (+10 to +1885) with GFP and expressed it in the pDXA vector under the actin15 promoter in bzpF– mutants. (B) Quantitation of bzpF expression: We measured the abundance of bzpF mRNA in AX4, bzpF–, and two independent ectopic expression transformants (BzpFGFPoe) by qRT-PCR in vegetative cells (0 hour, white bars) and in developing cells (20 hour, black bars). The data (arbitrary units, y-axis) are the means and standard errors of five replicates. (C) Partially penetrant expression of BzpF-GFP: We determined the fraction of GFP positive spores (%, y-axis) by flow cytometry of the ectopic expression strains and of a control strain expressing GFP alone (x-axis). (D) Complementation of the compromised spore phenotype in spores that express bzpF-GFP: We stained spores of the indicated strains (x-axis) with PI and used flow cytometry to determine the proportion (%, y-axis) of PI-positive (compromised) spores among the wild type (AX4) and among the GFP-positive spores of the transgenic strains. (E) Lack of complementation in spores that do not express BzpF-GFP: We stained spores of the indicated strains (x-axis) with PI and used flow cytometry to determine the proportion (%, y-axis) of PI-positive (compromised) spores among the parental (bzpF–) and among the GFP-negative spores of the transgenic strains. (F) The fruiting body morphology of the bzpFoe strains: Filter development of two bzpF-overexpressor strains (#1 and #2) showing morphologies that are nearly identical to the bzpF– phenotypes. Bar = 0.5 mm.