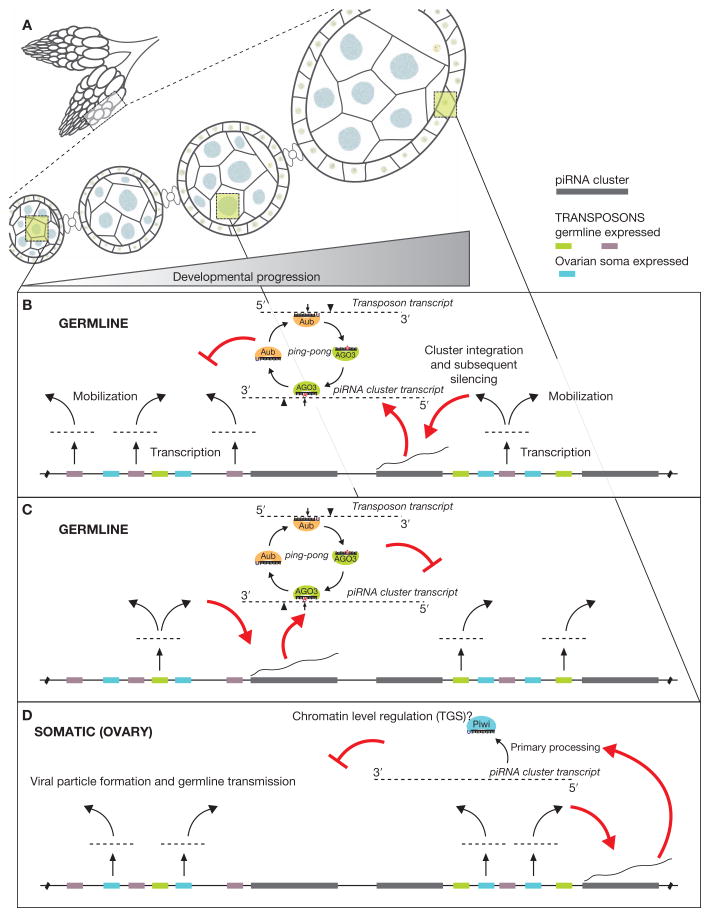
Figure 3.
Model for the acquisition of piRNA cluster–based transposon control. Tissue and developmental expression of piRNA clusters could act to combat the diverse expression programs of the myriad transposons in Drosophila. (A) The Drosophila ovary is a complex tissue, representing a full spectrum of oogenesis developmental stages and containing a mixture of both germline and somatic tissues. (B) Germline-expressed transposons (here, purple) eventually mobilize into a coexpressed piRNA cluster (red arrows). This mobilization leads to the entrance of that element into a potent silencing program involving piRNA “ping-pong” and leading to the PTGS (posttranscriptional gene silencing) of active transposon transcripts. (C) Same as B, only now a different set of transposons (green), expressed at a distinct developmental stage, are trapped into the piRNA pathway by a separate, yet coexpressed, piRNA cluster. (D) A somatic expressed cluster (flamenco) captures transposing gypsy family elements, which are transcriptionally active in somatic cells and possess the ability to package into viral particles that infect germline nuclei. However, in this case, transposition into the flameco cluster would need to occur and be selected for within the germline cells.