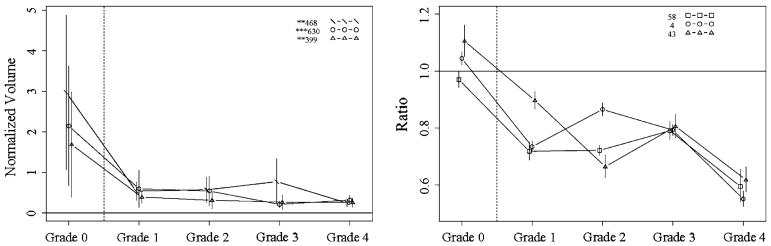
Fig. 2.
Protein profiles across fibrosis grades 0–4, showing the top candidates for discriminating grade 0 from grades 1–4, with DIGE (left) and iTRAQ (right). Proteins shown: DIGE spots 468 (complement factor H, haptoglobin, serum amyloid P-component), 630 (apolipoprotein A-I, complement C3, Ig kappa chain C region, tetranectin), and 399 (haptoglobin, serum amyloid P-component, kininogen-1, dermcidin); iTRAQ proteins 58 (complement component 8), 4 (complement component 4A), and 43 (insulin-like growth factor binding protein). Vertical bars indicate 95% confidence intervals