Abstract
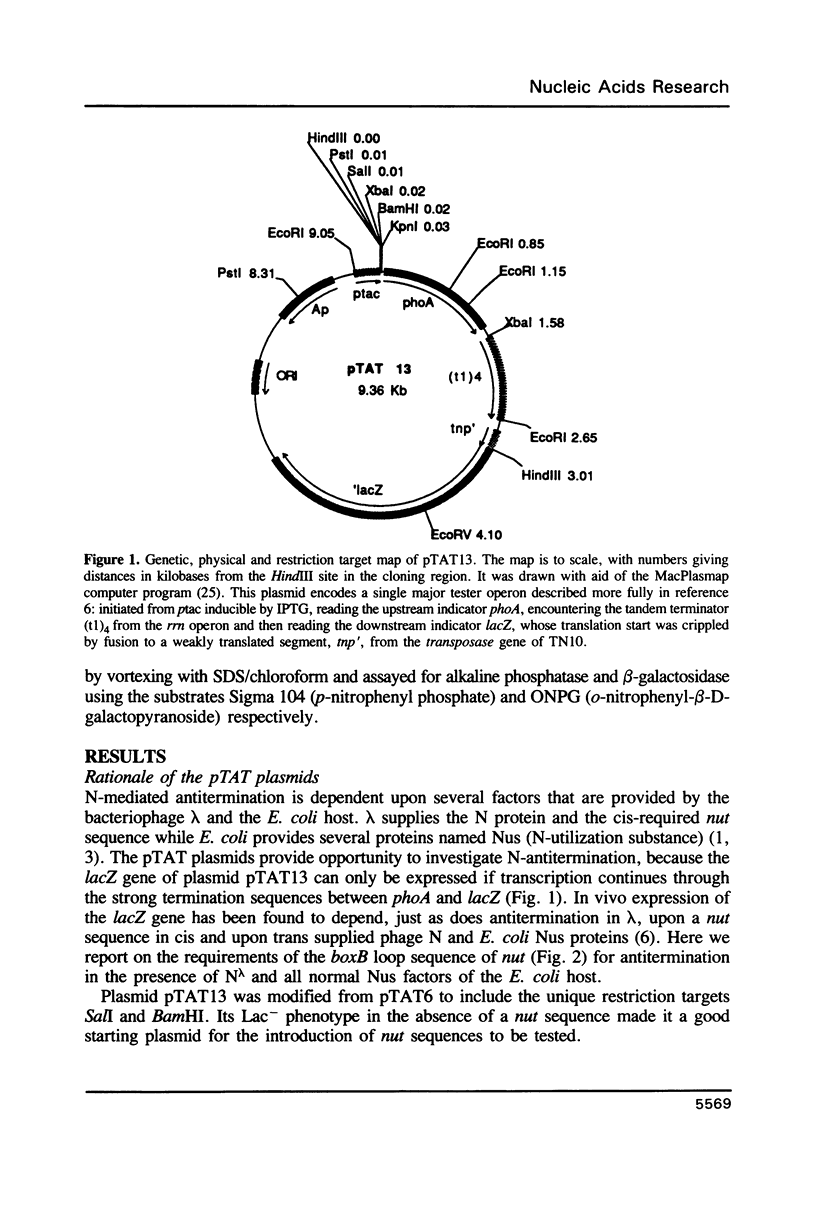
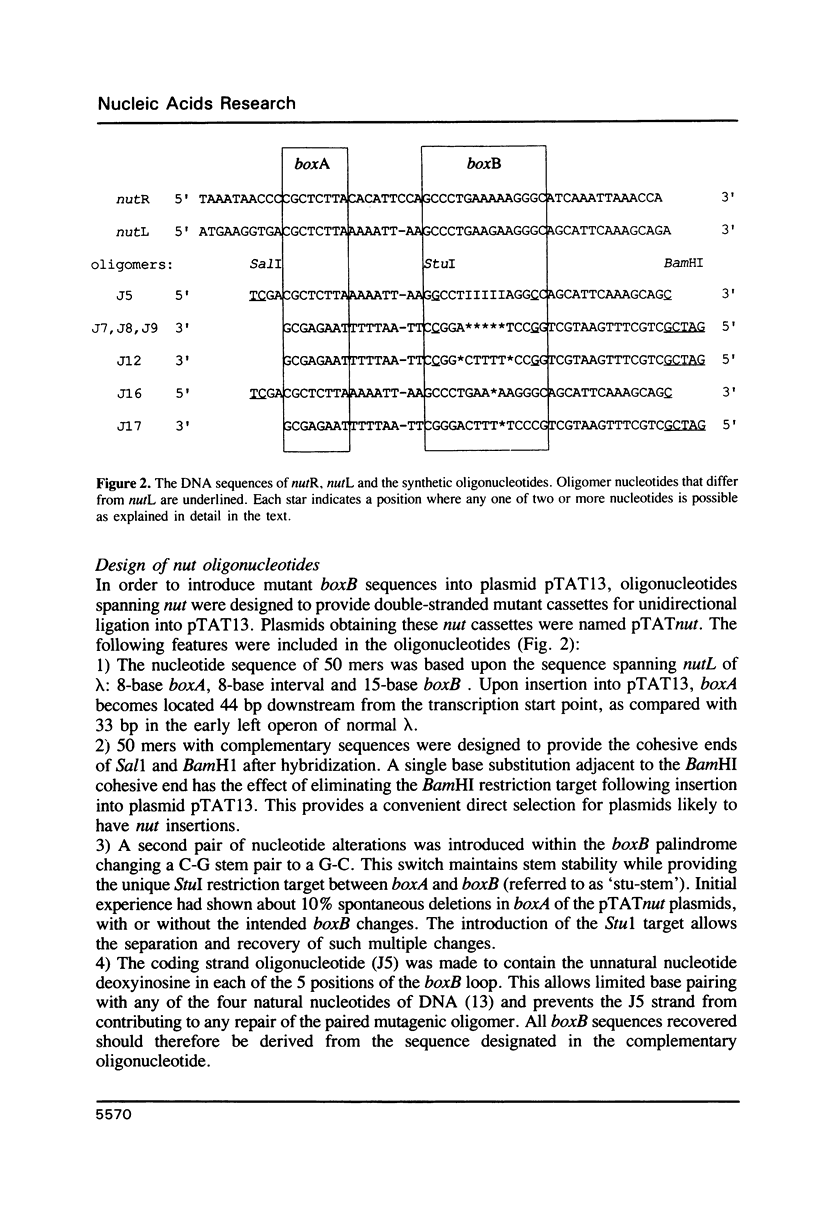
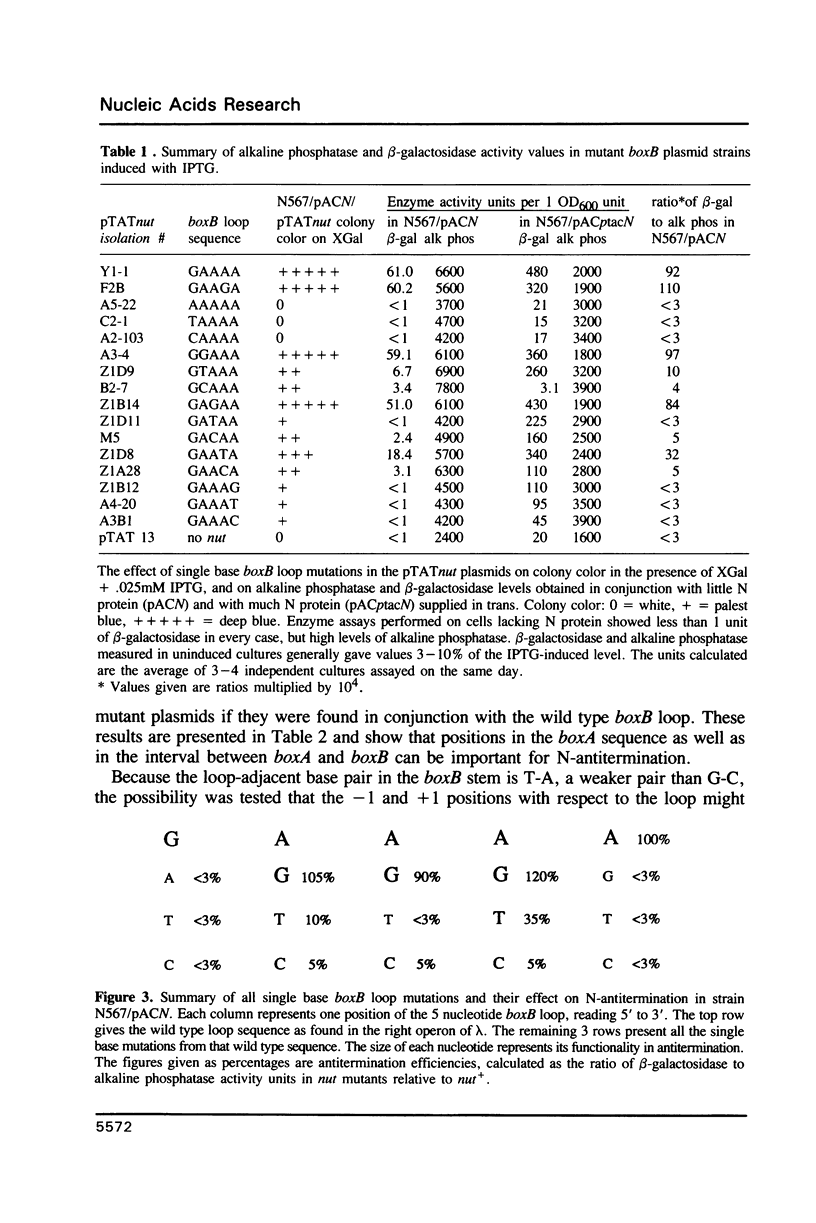
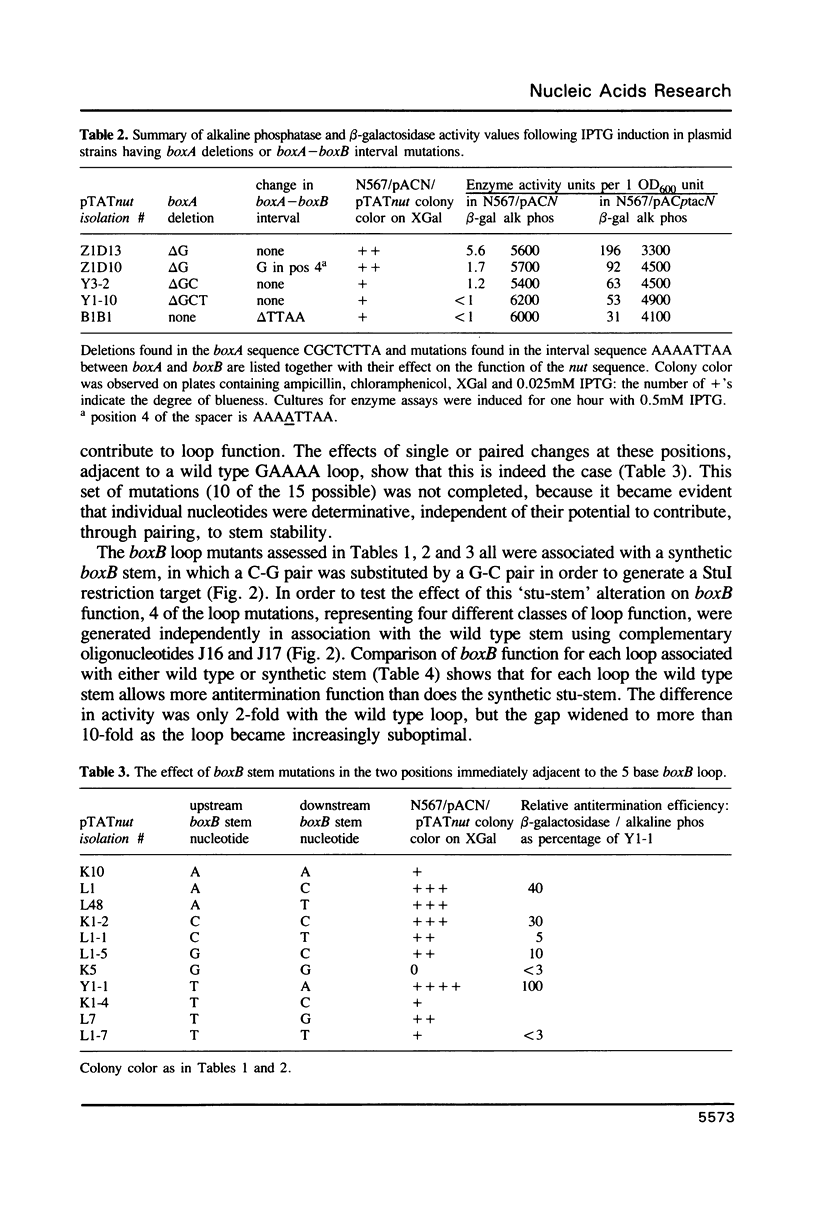
The 'N' antitermination proteins of lambdoid bacteriophages are essential for overcoming multiple transcription terminators located within the major early operons of these phages (1). In order for N proteins to function, a genome sequence specifying N utilization, nut, must be located within an operon, between the promoter and the terminators (2). Two components have been identified within nut: 8-base boxA, conserved among different phages and implicated in the recognition of host NusA protein, required for N function (3); 15-base boxB, an interrupted palindrome (4), diverged in sequence among different lambdoid phages and hypothesized to be the site of recognition for different N proteins, also diverged in sequence (5). Here we apply a plasmid for testing termination and antitermination of transcription (6) to identify mutations at all positions in the 5-7 base loop of lambda's boxB. Almost every base change at any position within the 5-7 base boxB loop was found to constrain antitermination of transcription by the N protein of bacteriophage lambda. These observations extend previous mutational knowledge of nut (7) and are consistant with the hypothesis that the boxB loop is the direct site of recognition for N protein. Variations among the effects of different base changes suggest differential contacts between N protein and bases of the boxB loop, whether in DNA or RNA.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J., Cameron V., de Haseth P. L., Uhlenbeck O. C. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983 May 24;22(11):2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin N. C. A plasmid to visualize and assay termination and antitermination of transcription in Escherichia coli. Plasmid. 1989 Jan;21(1):31–42. doi: 10.1016/0147-619x(89)90084-x. [DOI] [PubMed] [Google Scholar]
- Franklin N. C. Conservation of genome form but not sequence in the transcription antitermination determinants of bacteriophages lambda, phi 21 and P22. J Mol Biol. 1985 Jan 5;181(1):75–84. doi: 10.1016/0022-2836(85)90325-0. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Gottesman M., Shaw J. E., Pearson M. L. Protein degradation in E. coli: the lon mutation and bacteriophage lambda N and cII protein stability. Cell. 1981 Apr;24(1):225–233. doi: 10.1016/0092-8674(81)90518-3. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Masukata H., Tomizawa J. Transcriptional regulation of early functions of bacteriophage phi 80. J Mol Biol. 1988 Aug 5;202(3):551–563. doi: 10.1016/0022-2836(88)90285-9. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Echols H. The role of the N gene of phage lambda in the synthesis of two phage-specific proteins. Proc Natl Acad Sci U S A. 1968 Jun;60(2):707–712. doi: 10.1073/pnas.60.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidhaar-Olson J. F., Sauer R. T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science. 1988 Jul 1;241(4861):53–57. doi: 10.1126/science.3388019. [DOI] [PubMed] [Google Scholar]
- Salstrom J. S., Szybalski W. Coliphage lambdanutL-: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol. 1978 Sep 5;124(1):195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- Schauer A. T., Carver D. L., Bigelow B., Baron L. S., Friedman D. I. lambda N antitermination system: functional analysis of phage interactions with the host NusA protein. J Mol Biol. 1987 Apr 20;194(4):679–690. doi: 10.1016/0022-2836(87)90245-2. [DOI] [PubMed] [Google Scholar]
- Shortle D., Koshland D., Weinstock G. M., Botstein D. Segment-directed mutagenesis: construction in vitro of point mutations limited to a small predetermined region of a circular DNA molecule. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5375–5379. doi: 10.1073/pnas.77.9.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steege D. A., Cone K. C., Queen C., Rosenberg M. Bacteriophage lambda N gene leader RNA. RNA processing and translational initiation signals. J Biol Chem. 1987 Dec 25;262(36):17651–17658. [PubMed] [Google Scholar]
- Tanaka S., Matsushiro A. Characterization and sequencing of the region containing gene N, the nutL site and tL1 terminator of bacteriophage phi 80. Gene. 1985;38(1-3):119–129. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


