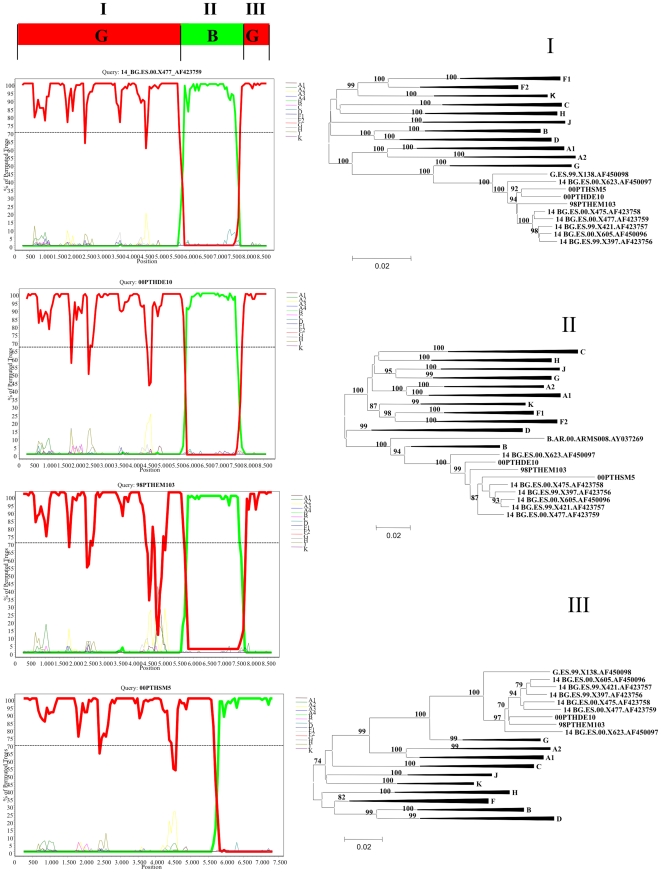
Figure 2. Bootscanning analysis of full-length genomes from Portuguese and Spanish CRF14_BG isolates.
The dashed line indicates the cut-off of 70%. The maximum likelihood phylogenetic trees were constructed with reference sequences from all HIV-1 subtypes. The bootstrap values supporting the internal branches defining a subtype or a CRF are shown. Bootstrap values of 70% or greater provide reasonable confidence for assignment of an individual segment to one or the other genotype. The scale represents number of base substitutions per site.