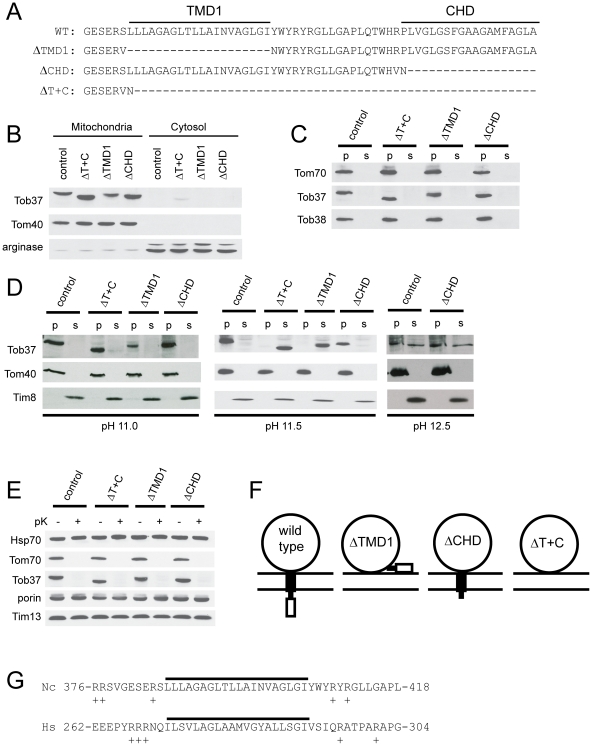
Figure 5. Role of predicted TMDs of Tob37.
(a) The WT row shows the sequence of the 63 amino acids at the C-terminus of wild type N. crassa Tob37. ΔTMD1 is the deletion constructed for the first possible transmembrane domain and is the name of the strain expressing this form of Tob37. Similarly for CHD, the second possible TMD found at the C-terminus of the protein, and for ΔT+C, the deletion of the last 56 amino acids of the protein which removes both possible TMDs. (b) Mitochondria and post-mitochondrial supernatants (cytosol) were isolated from strains expressing the mutant forms of Tob37 described in panel A. Samples of each were subjected to SDS-PAGE and transferred to nitrocellulose. The membrane was immunodecorated with the antibodies indicated on the left. The control was strain 76-26. Arginase represents a cytosolically localized control protein that is synthesized from two different start codons of the same locus [67] so that two bands of 41 kDa and 36 kDa are observed. (c) As in panel A except isolated mitochondria were treated for 30 min on ice with isolation buffer containing 0.5 M NaCl. Following the incubation period, mitochondria were pelleted. The supernatant was collected and desalted. The mitochondria were washed in isolation buffer and pelleted. Pelleted mitochondria and the desalted supernatant were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose and the membrane was probed with the antibodies indicated on the left. (d) Mitochondria from the strains indicated above the panel were subjected to alkali treatment using 0.1 M sodium carbonate at pH 11.0, 11.5, and 12.5 (indicated below each panel) as described in the legend to Figure 4B. (e) Mitochondria were isolated from each of the Tob37 deletion protein strains and treated with proteinase K as described in the Materials and Methods and Figure 4A. (f) The large circle represents the cytosolic domain of Tob37, the filled box is TMD1, and the open box is the CHD. Two horizontal lines represent the mitochondrial outer membrane. The predicted arrangement of the domains for wild type and each of the TMD/CHD deletions is indicated. (g) Comparison of potential tail-anchoring sequences of N. crassa (Nc) Tob37 and H. sapiens (Hs) Mtx1. The potential TMD is indicated by the solid line. The position of the region within each protein is indicated by the numbers flanking each amino acid sequence. The overall length of the N. crassa protein is 442 residues. The H. sapiens protein is 304 residues. Positively charged residues in the immediate flanking regions are indicated by the plus sign.