Figure 5.
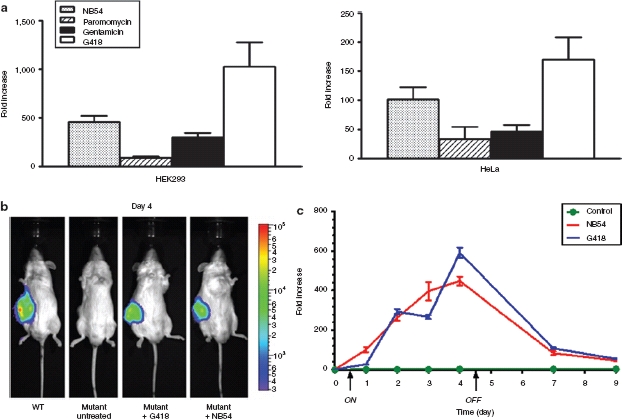
In vitro and in vivo regulation of hGLuc expression with the less toxic novel aminoglycoside NB54 (a) Fold induction of luciferase expression by various aminoglycosides in HEK293 and HeLa cells transduced with the HIV-2 lentiviral vector harboring the mutant hGLuc construct. 1 × 104 cells seeded in 96-well plates were exposed to NB54 (1.0 mg/ml), paromomycin (1.0 mg/ml), gentamicin (1.0 mg/ml), and G418 (0.25 mg/ml) for 48 hours before total samples from cultured media were assayed for luciferase activity. Data was represented as fold induction relative to untreated cells. All luciferase activity experiments were done in triplicate and repeated at least twice. (b) Noninvasive bioluminescence imaging of G418 or NB54 induced luciferase expression from subcutaneously (s.c.) implanted HEK293 xenograft mice on day 4. Control and mutants represent NOD-scid IL2Rgammanull (NSG) mice injected s.c. with transduced HEK293 cells harboring the mutant hGLuc construct exposed to phosphate buffered saline, G418 and NB54, respectively. Wild type (WT) represent NSG mice injected s.c. with transduced HEK293 cells harboring the wild-type hGLuc construct. Experiments were repeated twice and each time involved the use of at least three mice per experimental group. (c) The “On” and “Off” regulation of hGLuc expression in HEK293 xenograft model. Luciferase activity was measured in relative light units (RLU) from blood serum. The graph is represented in fold induction relative to luciferase reading before drug exposure (day 0). The “ON” and “OFF” arrows on the x-axis indicate when the drugs were administered and discontinued, respectively. Blood was collected, as described in the Material and Methods, prior to every injection of G418 and NB54. Luciferase activity experiments from blood serum were done in triplicate. Error bars represent standard deviation.
