Abstract
It remains unclear how speciation history might contribute to species-specific variation and affect species delimitation. We examined concordance between cytoplasmic genetic variation and morphological taxonomy in two fir species, Abies chensiensis and A. fargesii, with overlapping distributions in central China. Range-wide genetic variation was investigated using mitochondrial (mt) and plastid (pt) DNA sequences, which contrast in their rates of gene flow. Four mtDNA haplotypes were recovered and showed no obvious species' bias in terms of relative frequency. In contrast, a high level of ptDNA variation was recorded in both species with 3 common ptDNA haplotypes shared between them and 21 rare ptDNA haplotypes specific to one or other species. We argue that the lack of concordance between morphological and molecular variation between the two fir species most likely reflects extensive ancestral polymorphism sharing for both forms of cytoplasmic DNA variation. It is feasible that a relatively fast mutation rate for ptDNA contributed to the production of many species-specific ptDNA haplotypes, which remained rare due to insufficient time passing for their spread and fixation in either species, despite high levels of intraspecific ptDNA gene flow. Our phylogeographic analyses further suggest that polymorphisms in both organelle genomes most likely originated during and following glacial intervals preceding the last glacial maximum, when species distributions became fragmented into several refugia and then expanded in range across central China.
Keywords: Abies chensiensis, A. fargesii, biogeography, gene flow, mutation rate, species delimitation
Introduction
The repeated shifts in distribution ranges of plant and animal species caused by the Quaternary climatic oscillations have left distinct imprints on their current genetic structures (Hewitt, 2000). Thus, in addition to palaeobotanical data, the present-day distribution of molecular variation within plant species can provide evidence of species or vegetation shifts within a local region during the Pleistocene (for example, see Abbott et al., 2000; Petit et al., 2003). However, resolving the evolutionary history of genetic variation within and between closely related species is often challenging. On the one hand, an allopatric distribution across glacial refugia might promote interspecific genetic divergence through fixing species-specific haplotypes via genetic drift and selection (Avise and Walker, 1998). On the other hand, secondary contact following range expansions of previously isolated species can result in genetic changes through introgression (McKinnon et al., 2004).
DNA sequences are employed increasingly to examine concordance between genetic variation and traditional morphological boundaries among closely related species (for example, see Jaramillo-Correa et al., 2008; Du et al., 2009; Zhou et al., 2010). Such studies suggest that molecular polymorphisms of neutral markers are often widely shared between closely related species, and that fixed genetic differences vary according to the genomes or genes studied (see review by Petit and Excoffier, 2009). In conifer species, plastid (pt) DNA variation, which is mainly paternally inherited and shows high rates of gene flow, is often more species specific than mitochondrial (mt) DNA variation, which is mainly maternally inherited and exhibits lower rates of gene flow (Du et al., 2009; Zhou et al., 2010). Three possible mechanisms may account for this difference, which are often difficult to distinguish in empirical studies. First, high rates of intraspecific gene flow may override introgression and produce more diagnostic genetic differences between species (Petit and Excoffier, 2009), as suggested by recent simulation studies (Currat et al., 2008). Second, high rates of intraspecific gene flow may promote rapid lineage sorting of ancestral polymorphisms, such that alleles exhibiting higher dispersal may rapidly become more species specific (Hoelzer, 1997; Zhou et al., 2010). Finally, mutation-rate heterogeneity between different DNAs (Wolfe et al., 1987) could increase the development of species-specific genotypes in isolation (Sloan et al., 2008). A further complication is that ‘speciation genes' might spread rapidly across populations of two diverging species due to powerful selection, causing reproductive isolation and morphological delimitation to occur (for example, see Ting et al., 2000) before sufficient time has passed to achieve monophyly of neutral makers (de Queiroz, 2007). Thus, a genetic mosaic might continue to persist for a long time across two species with high genetic divergence between them occurring in and around ‘speciation genes', whereas low divergence prevails at unlinked or loosely linked neutral markers (Via, 2009). It is evident, therefore, that the sharing and partitioning of neutral genetic variations between species will depend greatly on historical processes occurring during and following their origin, that is, the speciation history (Wiens, 2007).
Here, we report a case study of genetic divergence between two fir species, Abies chensiensis Tieghem and A. fargesii Franchet, on the basis of surveys of mt and ptDNA sequence variation. These two species occur exclusively in central China where many indigenous species shifted their distribution ranges in response to Quaternary climatic oscillations even though the region was never covered by ice sheets during the Quaternary (Chen et al., 2008; Tian et al., 2009). The occurrence of range fragmentation and adaptation to new niches during this period may have promoted speciation in the region (Qian and Ricklefs, 2000). A. chensiensis is an endangered plant that is usually found scattered in small forest fragments at elevations from 2300 to 3000 m, whereas A. fargesii is a dominant member of coniferous forests found at elevations between 1500 and 3900 m (Fu et al., 1999). Morphologically, the two species are distinguished by A. chensiensis having branchlets that are yellowish gray or brownish yellow, mature seed cones that are light brown, inserted bracts, and pale-green leaf needles that are 3.9–4.6 cm long and 2.1–2.6 mm wide, whereas A. fargesii has purplish brown or reddish brown branchlets, dark purple or reddish brown mature seed cones, exserted bracts with much longer cusps, and dark-green leaf needles that are 1.4–1.8 cm long and 1.4–2.0 mm wide (Fu et al., 1999). As in other fir species, the pollen of both species is wind-dispersed and seeds are dispersed by animals (Fu et al., 1999). On the basis of their differences, A. chensiensis and A. fargesii are often grouped into different sections of genus Abies (Liu, 1971; Farjon and Rushforth, 1989). Their exact phylogenetic relationship remains uncertain, although our unpublished phylogenetic analysis of all fir species in China using ptDNA sequence variation suggests that they are closely related, and most likely sister species.
The biogeography and delimitation of fir species outside China have been explored previously using mtDNA and ptDNA markers (for example, see Tsumura and Suyama, 1998; Liepelt et al., 2002, 2010; Jaramillo-Correa et al., 2008). These studies have revealed different phylogeographic structures and/or inconsistent patterns of species delimitation for the two markers, with mtDNA variation tending to reveal stronger intraspecific genetic structure and weaker resolution of species boundaries when compared with ptDNA. In this study, we examined the genetic structure and evolutionary relationships among a total of 254 individuals from 25 populations of A. chensiensis and A. fargesii across their overlapping and allopatric ranges. In doing so, we aimed to address the following questions: (1) how is ptDNA and mtDNA variation distributed within and between the two species? Is ptDNA variation more species specific than mtDNA variation? If so, is this likely to result from reduced introgression or rapid lineage sorting of ptDNA due to a higher rate of intraspecific gene flow of ptDNA, or is it more likely to stem from a fast mutation rate of this genome? (2) Are the biogeographic histories of these two high-altitude tree species consistent with those reported for other species in the region?
Materials and methods
Species and population sampling
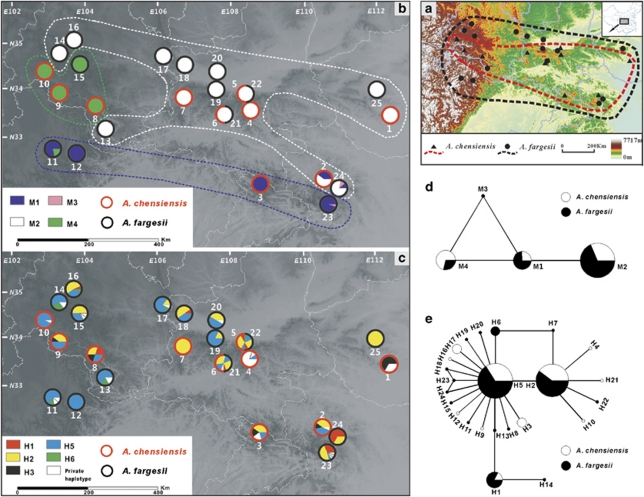
We sampled morphologically pure and autochthonous populations of A. chensiensis and A. fargesii from almost the entire range of the two species in China, such that a total of 254 individuals from 25 populations were sampled (Figure 1a; Supplementary Table S1). At most localities, only one species was present (Figure 1b and c). However, at two localities, both species occurred parapatrically (populations 5 and 22 were parapatric at one site, whereas populations 6 and 21 were parapatric at another site). At these two localities, A. chensiensis was particularly abundant at lower elevations, whereas A. fargesii was mainly restricted to higher altitudes (above 2200 m). At altitudes between 2000 m and 2200 m, some trees were morphologically intermediate and possibly of hybrid type. Hence, care was taken at these localities to collect individuals of different species separated by a minimum of 200 m in altitude to avoid sampling possible hybrids. Fresh leaves of 1–19 individuals, spaced at least 100 m apart, were collected from each population and dried in silica gel. For the phylogenetic ptDNA analysis (see below), A. nordmanniana was selected as outgroup on the basis of a previous molecular study showing its sister-group relationship with two studied species (Xiang et al., 2009). Voucher specimens of representatives of all populations of the two species and the outgroup species were collected and stored in the herbarium of Lanzhou university (more details in Supplementary Table S1). We recorded the latitude, longitude and altitude of each location site by Extrex GIS monitor (Garmin, Taiwan).
Figure 1.
(a) Sampling localities (details in Supplementary Table S1) and distributions of A. chensiensis (marked by black solid triangle and red dashed line) and A. fargesii (marked by black solid circle and black dashed line) in central China. (b) Geographical distribution of four mtDNA haplotypes (mitotypes) detected in A. chensiensis (marked by the red circle lines) and A. fargesii (marked by the black circle lines). Locations where both species were collected, but from different altitudes, are represented by circles that are half red and half black. (c) Geographical distribution of main and private ptDNA haplotypes in these two species. All haplotypes that are found in more than one population are color-coded, whereas private haplotypes are always represented by white circles. (d, e) Haplotype network of 4 mitotypes and 24 ptDNA haplotypes, identified in this study. Each mitotype or ptDNA haplotype is represented by a circle whose size is proportional to its frequency over all populations. Lines between haplotypes indicate one mutational change. The white and black segments within circles in (d) and (e) represent the relative proportions of each mitotype and ptDNA haplotype in A. chensiensis and A. fargesii, respectively.
DNA extraction, amplification and sequencing
Total genomic DNA was extracted from approximately 20 mg of silica gel-dried, leaf needle material per sample, according to a cetytrimethyl ammonium bromide procedure (Doyle and Doyle, 1987). Variation was determined for the mtDNA sequence spanning intron 4 of subunit 5 of the mt NADH dehydrogenase gene (nad5 intron 4) and intron 1 of subunit 7 of the same gene (nad7 intron 1), described by Wu et al. (1998) and Jaramillo-Correa et al. (2004). In addition, ptDNA variation was determined for the trnS–trnG and trnL–trnF fragments following Demesure et al. (1995) and Taberlet et al. (1991). PCR was carried out in a 25-μl volume, containing 10–40 ng plant DNA, 50 m Tris-HCI, 1.5 m MgCl2, 250 μg ml−1 bovine serum albumin, 0.5 m dNTPs, 2 μ of each primer and 0.75 U of Taq polymerase. All reactions were performed with the following program: 4 min at 94 °C, 37 cycles of 40 s at 94 °C, 40 s of annealing at 58 °C for all the mt genes and 60 °C for pt genes, 1.20 min at 72 °C, with a final 7-min extension at 72 °C, and reactions were held at 4 °C until further processing.
We then purified PCR products using a TIANquick Midi Purification Kit following the recommended protocol (TIANGEN, Beijing, China). Sequencing reactions were performed with the PCR primers described above and internal primers for ptDNA trnL–trnF (primer ‘e': 5′-GGTTCAAGTCCCTCTATCCC-3′) using ABI Prism Bigdye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Forward, reverse or internal sequences were checked carefully by eye for all new mutations. Depending on the quality of sequence and the site of mutation, the forward, the reverse or the internal primers were then used for large-scale sequencing. We aligned DNA sequences with CLUSTAL X version 1.83 (Thompson et al., 1997) and refined the alignments by hand. A matrix of combined sequences was constructed for the 254 individuals examined, in which 4 different mtDNA sequences (mitotypes) and 24 different ptDNA sequences (ptDNA haplotypes) were identified. These mt and pt fragment sequences have been deposited in GenBank under accession numbers: HM851212-HM851248.
Data analysis
We first calculated haplotype gene diversity (h) and nucleotide diversity (π) of mtDNA and ptDNA for each population (hS, πS) and each species (hT, πT) using DnaSP version 5.0 (Librado and Rozas, 2009). To determine phylogenetic relationships among mtDNA and ptDNA haplotypes, we constructed median-joining networks (Bandelt et al., 1999) with NETWORK version 4.5.1.0 (available at http://www.fluxus-engineering.com). Furthermore, we also used PAUP* 4.0b10 (Swofford, 2002) to perform maximum parsimony analyses on ptDNA haplotypes, using A. nordmanniana as outgroup. Maximum parsimony analyses involved a heuristic search strategy with 100 replicates of random addition of sequences, in combination with ACCTRAN character optimization, MULPARS+TBR (tree bisection–reconnection) branch swapping and STEEPEST DESCENT options on. The robustness of maximum parsimony trees was estimated by 1000 bootstrap replicates. Bayesian inference was conducted using MrBayes version 3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003), with the best-fitting TrN+I model selected by Modeltest version 3.06 (Posada and Crandall, 1998). Four chains of the Markov chain Monte Carto were run each for 1 000 000 generations retaining every 100th sample, after discarding a burn-in of 30 000 generations. Each insertion/deletion (indel) was considered as a single mutation event, and all indels were therefore coded as single positions in the final alignment. In cases where it was not possible to code the indel variation by a single alignment position (overlapping indels), modified complex indel coding was used to code indels (Simmons and Ochoterena, 2000).
We used SAMOVA (spatial analysis of molecular variation; Dupanloup et al., 2002) to explore the population structure of sampling areas across the range of A. chensiensis and A. fargesii, without a priori hypotheses of the expected structure. This method was used to identify clusters of populations that are geographically homogeneous and genetically differentiated from each other. A simulated annealing process was run to obtain the number of K groups that maximizes FCT (proportion of total genetic variance due to differences among groups of populations). For these analyses, we used 100 simulated annealing processes for each value of K from K=2 to K=8 for both mtDNA and ptDNA haplotypes, and chose either the number of groups (K) that gave the highest FCT or the number at which FCT began to plateau.
We also calculated average gene diversity within populations (HS), total gene diversity (HT) and the coefficients of differentiation GST and NST for each species, separately and combined, for both mtDNA and ptDNA markers, using PERMUT (available at http://www.pierroton.inra.fr/genetics/labo/ Software/Permut/). In such analyses, GST considers only haplotype frequencies, whereas NST takes into account both haplotype frequencies and genetic similarity between haplotypes. Phylogeographic structure was inferred by testing whether NST was significantly larger than GST using a permutation test with 1000 random permutations of haplotypes across populations (Pons and Petit, 1996). Taking the different levels of variation obtained from mtDNA and ptDNA markers into account, standardized GST values (G′ST) following Hedrick (2005) were calculated using RecodeData version 0.1 (Meirmans, 2006), in conjunction with FSTAT (Goudet, 2001). Hierarchical partitioning of diversity among species, populations and individuals was estimated on the basis of AMOVA(Excoffier et al., 1992), which considered both haplotype frequency differences and the number of site differences among observed haplotypes using the program Arlequin version 3.5 (Excoffier and Lischer, 2010), with significance tested by a non-parametric permutation procedure with 1000 permutations. A Mantel test assessed the significance of isolation by distance between populations with 1000 random permutations on matrices of pairwise population FST values and the natural logarithm of geographical distances (Rousset, 1997).
In an attempt to infer demographic processes, a mismatch distribution analysis was carried out for each species, with each phylogeographic group being arranged by SAMOVA partitions and all individuals using Arlequin version 3.5 (Excoffier and Lischer, 2010), separately. The shape of the graph of the mismatch distribution is expected to be multimodal in samples drawn from populations at demographic equilibrium, whereas unimodal distributions are generally found in populations that have passed through a recent demographic expansion (Rogers and Harpending, 1992). The sum of squared deviations and raggedness indices (r) between observed and expected mismatch distributions were used as test statistics; their P-values represented the probability of obtaining a simulated sum of squared deviations greater than or equal to the one observed. We also performed Fu's FS test and calculated Tajima's D to confirm evidence of demographic expansion. Estimation and testing were done with Arlequin version 3.5 (Excoffier and Lischer, 2010) using 10 000 bootstrap replicates for both Fu's FS and Tajima's D. Finally, separate haplotype networks for each species and phylogeographic group were constructed using median-joining networks with NETWORK version 4.5.1.0 (Bandelt et al., 1999; available at http://www.fluxus-engineering.com) to obtain additional information about the past demographic history of the two species.
If the hypothesis of rapid expansion were not rejected, the relationship τ=2ut (Rogers and Harpending, 1992) was used to estimate the age of expansion (t), where u is the mutation rate for the DNA sequences. The value for u was calculated as u=μkg, where μ is the substitution rate per nucleotide site per year (s s−1 y−1), k is the average sequence length of the DNA region under study, and g is the generation time in years. We estimated expansions using μ=2.61 × 10−10–4.02 × 10−10 s s−1 y−1, which represent a range of neutral substitution rates for ptDNA in Pinaceae on the basis of different fossil calibrations (Gernandt et al., 2008). This produced a range of values for u and hence also for t. One caveat is that a slight bias of nucleotide substitution rates for ptDNA may exist in Abies. Thus, the substitution rates used here are only approximate estimates, although they are consistent with estimates reported recently for other conifer species (Willyard et al., 2007; Gugger et al., 2010). An average haplotype sequence length of 1111 bp and a minimum estimate of generation time of 25 years in Pinaceae were used in the calculations (Brown et al., 2004). Times of expansion were calculated separately for each species, phylogeographic group and all individuals.
Results
mtDNA variation
Polymorphism was observed within both mtDNA regions examined: nad5 intron 4 and nad7 intron 1. Different repeats of a 4-bp microsatellite formed three different size variants in intron 4 of nad5, whereas polymorphism in intron 1 of nad7 was produced by a single insertion at position 228 (Supplementary Table S2). When combined, these sequences resulted in identification of four distinct mitotypes ranging in size from 1033 to 1041 bp, with an alignment length of 1045 bp, and distributed as shown in Figure 1b and Supplementary Table S1. The three most frequent mitotypes (M1, M2 and M4) were shared between both species, whereas mitotype M3 was specific to A. fargesii, where it occurred at low frequency in populations 23 and 24 (Figure 1b and d). In all, 21 of the 25 populations surveyed were fixed for a single haplotype, whereas the remaining four polymorphic populations harbored two or three haplotypes. In the mitotype network, the four mitotypes were separated by only one or two mutational changes and, hence, were closely related (Figure 1d).
Estimates of h and π of mtDNA for each population are summarized in Supplementary Table S1. The analysis of spatial genetic structure for mitotype variation using SAMOVA showed that the value of FCT reached a plateau when the K equaled 3 (FCT=0.920). For K⩾3, some groups comprised a single population, indicating the disappearance of geographical structure (Supplementary Table S3). One of the three groups, termed the north-central group, was characterized by a high frequency of mitotype M2 and comprised populations 1–2, 4–7, 13–14, 16–22, 24. A second group, termed the western group, comprised populations 8–10 and 15, which were fixed for mitotype M4. The remaining group, termed the southern group, comprised populations 3, 11, 12 and 23, and was characterized by a high frequency of mitotype M1. Each group exhibited a distinct geographical range that neither overlapped those of other groups nor coincided with taxon identification (Figure 1b).
HT on the basis of mitotype variation across both species was much higher than HS, and consequently, both GST and NST were high in the two species (Table 1). There was also significant phylogeographic structure when populations from both species were considered together (NST=0.934>GST=0.905; P<0.05, Table 1). The G′ST for mtDNA variation was slightly higher than GST for both species (Table 1), whereas AMOVA showed that the percentage of variation explained at each hierarchical level was nearly identical across species (Table 2). Only 1.19% of the total mtDNA variation was attributed to variation between species, whereas 87.81% was attributed to variation among populations within species (Table 2). Mantel tests revealed a significant correlation between genetic divergence and geographical distance between populations combined over species (r=0.224, P<0.05) within A. chensiensis (r=0.521, P<0.05), but not in A. fargesii (r=0.130, P=0.179) (Table 1).
Table 1. Genetic diversity estimates and Mantel tests for mtDNA and ptDNA variations in Abies chensiensis and A. fargesii.
| Species | HS | HT | GST | NST | G′ST | Mantel test r (P) |
|---|---|---|---|---|---|---|
| mtDNA variation | ||||||
| A. chensiensis | 0.051 | 0.636 | 0.919 | 0.949* | 0.964 | 0.521 (0.006) |
| A. fargesii | 0.051 | 0.484 | 0.894 | 0.918, NS | 0.932 | 0.130 (0.179) |
| Total | 0.051 | 0.542 | 0.905 | 0.934* | 0.948 | 0.224 (0.011) |
| ptDNA variation | ||||||
| A. chensiensis | 0.544 | 0.780 | 0.303 | 0.300, NS | 0.663 | 0.087 (0.316) |
| A. fargesii | 0.565 | 0.694 | 0.185 | 0.169, NS | 0.418 | 0.324 (0.009) |
| Total | 0.556 | 0.737 | 0.245 | 0.233, NS | 0.537 | 0.158 (0.045) |
Abbreviations: GST, interpopulation differentiation; G′ST, the standardized GST; HS, average gene diversity within populations; HT, total gene diversity; mtDNA, mitochondrial DNA; ptDNA, plastid DNA; NST, the number of substitution types. *NST is significantly different from GST (P<0.05); NS indicates that NST is not significantly different from GST.
Table 2. AMOVA of mtDNA and ptDNA haplotype variation in Abies chensiensis and A. fargesii.
| Species | Source of variation | d.f. | SS | VC | V% | F-statistic |
|---|---|---|---|---|---|---|
| mtDNA haplotype | ||||||
| All | Among species | 1 | 3.214 | 0.003 | 1.19 | FCT=0.012 |
| Among populations within species | 23 | 56.956 | 0.244 | 87.81 | FSC=0.889* | |
| Within populations | 229 | 6.992 | 0.031 | 11.00 | FST=0.890* | |
| Total | 253 | 67.161 | 0.278 | |||
| A. chensiensis | Among populations | 9 | 24.202 | 0.280 | 86.99 | FST=0.870* |
| Within populations | 86 | 3.600 | 0.042 | 13.01 | ||
| Total | 95 | 27.802 | 0.322 | |||
| A. fargesii | Among populations | 14 | 32.754 | 0.223 | 90.37 | FST=0.904* |
| Within populations | 143 | 3.392 | 0.024 | 9.63 | ||
| Total | 157 | 36.146 | 0.246 | |||
| ptDNA haplotype | ||||||
| All | Among species | 1 | 0.835 | 0.003 | 0.79 | FCT=0.008 |
| Among populations within species | 23 | 24.412 | 0.078 | 21.70 | FSC=0.215* | |
| Within populations | 229 | 64.757 | 0.283 | 79.08 | FST=0.209* | |
| Total | 253 | 90.004 | 0.358 | |||
| A. chensiensis | Among populations | 9 | 12.264 | 0.117 | 31.17 | FST=0.312* |
| Within populations | 86 | 22.184 | 0.258 | 68.83 | ||
| Total | 95 | 34.448 | 0.375 | |||
| A. fargesii | Among populations | 14 | 12.149 | 0.055 | 15.54 | FST=0.155* |
| Within populations | 143 | 42.573 | 0.298 | 84.46 | ||
| Total | 157 | 54.722 | 0.353 |
Abbreviations: AMOVA, analysis of molecular variance; d.f., degrees of freedom; mtDNA, mitochondrial DNA; ptDNA, plastid DNA; SS, sum of squares; VC, variance component. *P<0.001, 1000 permutations.
ptDNA variation
In all, 5 indels and 13 substitutions were detected in the trnS–trnG and trnL–trnF fragments leading to the resolution of 24 different ptDNA haplotypes (H1–H24) among trees surveyed (Supplementary Table S2). Haplotype sizes ranged from 1083 to 1139 bp, with a sequence alignment of 1186bp. The three most frequent ptDNA haplotypes (H1, H2 and H5) were shared between species and located near the center of the haplotype network, while numerous closely related tip haplotypes that were rare and species specific were derived from each of these haplotypes (Figure 1c and e; Supplementary Table S1). Phylogenetic analysis of all ptDNA haplotypes, with A. nordmanniana used as outgroup, showed that phylogenetic relationships among ptDNA haplotypes were poorly resolved (Supplementary Figure S1).
Estimates of haplotype and nucleotide diversity within populations for ptDNA were much higher than those for mtDNA, but varied widely among populations in each species, although averages were similar for both species (Supplementary Table S1). SAMOVA failed to reveal any meaningful phylogeographic groups of ptDNA haplotypes (Supplementary Table S3). In contrast to mtDNA variation, HT on the basis of ptDNA haplotype variation was only slightly higher than HS for both species. This resulted in relatively lower values for GST and NST, and in neither species was NST significantly greater than GST (Table 1). However, GST for ptDNA variation might considerably underestimate the extent of population differentiation when there is high genetic variation within populations (Hedrick, 2005). This appears to be the case here as G′ST, which overcomes this problem, was indeed much higher than GST in both species (G′ST=0.663 in A. chensiensis, G′ST=0.418 in A. fargesii; Table 1). AMOVA confirmed that most ptDNA diversity was distributed within populations (79.08%), with negligible variation present between species (Table 2). A Mantel test showed that ptDNA variation was correlated with geographical distance across the distribution of both species (r=0.158, P<0.05) within A. fargesii (r=0.324, P<0.05), but not in A. chensiensis (r=0.087, P=0.316) (Table 1).
Historical demography on the basis of ptDNA sequence variation
The mismatch distributions for both fir species, for the three mitotype groups identified by SAMOVA, and for all individuals, showed a good fit to the expansion model with curves appearing to be unimodal (Supplementary Figure S2). In addition, the nonsignificant variance (sum of squared deviations) and raggedness index, plus the significant negative values for Fu's FS and Tajima's D in these samples, supported a model of sudden demographic expansion (Supplementary Table S4), as did the star-like patterns resolved in the ptDNA networks for each species and mtDNA group (Supplementary Figure S3). On the basis of recent fossil-calibrated neutral substitution rates for ptDNA observed in Pinaceae, we calculated the demographic expansions to have occurred between 42 000 and 105 000 years ago (kya), according to mismatch analysis (Rogers and Harpending, 1992) (Supplementary Table S4).
Discussion
In agreement with results from several previous studies on conifers (Liepelt et al., 2002; Du et al., 2009), we recorded low levels of mtDNA polymorphism in the two fir species, A. chensiensis and A. fargesii, which have overlapping distributions in the Tsinling and adjacent mountains of central China. Of the four mitotypes identified, three were shared between both species, with variation distributed mainly between populations rather than within populations or between species. In contrast, a high level of ptDNA polymorphism was recorded in both fir species, due largely to high levels of variation within rather than between populations. Neither mtDNA variation nor ptDNA variation delimited the two species from each other, despite the fact that rare ptDNA haplotypes were species specific. The difference in population structure exhibited by mtDNA relative to ptDNA variation most likely reflects a higher rate of sequence evolution for ptDNA (Wolfe et al., 1987).
Biogeographic history
Mitotype variation across both species showed significant phylogeographic structure (NST>GST, P<0.05) and SAMOVA-clustered populations into three mitotype groups. Each of these groups was characterized by a particular mitotype and exhibited a distinct geographical distribution; thus, a north-central group was characterized by a high frequency of mitotype M2, a western group was fixed for mitotype M4, and a southern group was characterized by a high frequency of mitotype M1. The occurrence of these groups suggests that at least three independent refugia were maintained for both species (or their common ancestor) possibly during Quaternary glacial periods when their distributions may have fragmented in central China. If this is correct, then the current distribution of populations is likely to have resulted from expansions from these different refugia, with secondary contact occurring in some areas giving rise to populations (for example, populations 2, 11, 23 and 24) polymorphic for mitotypes diagnostic of the different phylogeographic groups. Other surveys of molecular variation in different plant species occurring in central China, such as Taxus wallichiana (Gao et al., 2007), Pinus tabulaeformis (Chen et al., 2008), Ostryopsis davidiana (Tian et al., 2009), Eurycorymbus cavaleriei (Wang et al., 2009), have also indicated that multiple refugia existed in this region during the Quaternary.
Although SAMOVA did not detect a similar population structure for ptDNA variation, recent range expansions of the two fir species were also indicated by population genetic and phylogenetic analyses of ptDNA haplotype variation. Thus, the occurrence of significantly negative values for Fu's FS and Tajima's D, unimodal mismatch distributions, and ‘star-like' phylogenies of ptDNA haplotypes within and across species, indicated that rapid expansions took place in both species in the relatively recent past (Hwang et al., 2003). Importantly, these signatures of rapid expansion were evident when analyses were restricted to ptDNA haplotypes within population groups representing the three mtDNA groups detected by SAMOVA. This supports the hypothesis of rapid migrations from isolated refugia (see above).
The much higher level of ptDNA haplotype variation relative to mitotype variation recorded across both fir species might reflect a higher mutation rate for ptDNA (see below). A difference in mutation rate has been suggested to be a cause of two very different types of genetic composition for populations that have undergone a recent bottleneck, followed by rapid expansion. In such populations, a single haplotype is likely to be present for sequences exhibiting a low mutation rate, whereas a dominant haplotype with multiple rare alleles, as found for ptDNA haplotype variation in the present study, is more likely to be present for sequences showing a high mutation rate (for example, see Akey et al., 2004).
An indication of when rapid expansions from refugia were most likely to have occurred was obtained for both fir trees from mismatch analyses of ptDNA haplotype variation. These analyses suggested that the species underwent a notable range expansion, approximately 42–105 kya, that is following the penultimate glacial period that occurred in China between approximately 333 and 136 kya (Yi et al., 2005). This range expansion might reflect the most recent demographic shift of conifer forests in the Tsinling and adjacent mountains of central China in response to climatic oscillations during the Pleistocene (Zheng, 2000). Thus, unlike species in North America and Europe, which commonly expanded in range after the last glacial maximum (around 18 kya) (Abbott et al., 2000; Hewitt, 2000; Petit et al., 2003), our results suggested that the last glacial maximum had little impact on the demographic history of the two fir species we studied from central China. This accords with results obtained from recent studies on a few other plants and animals in Asia, such as Dysosma versipellis (Qiu et al., 2009), Allium przewalskianum (Wu et al., 2010) and Leucodioptron canorum (Li et al., 2009), all of which suggest that the last glacial maximum had a less important effect on the demographic histories of species in this region relative to earlier Pleistocene glaciations.
Cytoplasmic DNA variation and species delimitation
AMOVA showed that only ∼0.79% of total ptDNA variation and ∼1.19% of total mtDNA variation was partitioned between the two fir species. Although some ptDNA haplotypes were species specific, the three most common ptDNA haplotypes (H1, H2 and H5), along with the three common mitotypes (M1, M2 and M4), were widely shared across both species (Figure 1). Shared polymorphisms across closely related species appear to be common in fir, pine and spruce genera of the Pinaceae (for example, see Tsumura and Suyama, 1998; Godbout et al., 2008; Jaramillo-Correa et al., 2008; Du et al., 2009). Sharing might result from introgression following secondary contact between long diverged species or, alternatively, to the maintenance of ancestral polymorphisms due to incomplete lineage sorting within recently diverged species (Wiens, 2007; de Queiroz, 2007). Recently, it was shown that in most conifers, ptDNA delimits closely related species more effectively than does mtDNA (Du et al., 2009). This difference in behavior of the pt and mtDNA could be explained in part by differences in the level of intraspecific gene flow, that is, the more the gene flow within species, the less the introgression between species (Currat et al., 2008; Petit and Excoffier, 2009). If introgression were the main cause of sharing of haplotypes between species, sharing would be expected to be more evident between parapatric than allopatric populations (Palmé et al., 2004). However, in the present study, the amount of sharing of mitotypes and ptDNA haplotypes between species showed no bias with regard to either parapatric or allopatric population distributions. Although we cannot fully exclude the possibility that introgression contributed to the observed genetic patterns between the two species, the shallow genealogy and the relative interior position of shared haplotypes in the species networks for both mtDNA and ptDNA suggests that these species may have diverged too recently for ancestral polymorphisms to have been sorted to the stage of reciprocal monophyly (Crandall and Templeton, 1993). It is feasible, therefore, that incomplete sorting of ancestral polymorphisms is the more likely explanation for the sharing of haplotypes between the two fir species.
The finding that all species-specific ptDNA haplotypes originated between 42 and 105 kya indicates that the speciation events that gave rise to A. chensiensis and A. fargesii probably occurred very recently in the Quaternary, as suggested for many endemic species in eastern Asia (Qian and Ricklefs, 2000). It is also likely that the near lack of species-specific mitotypes might stem from a low mutation rate for mtDNA (Wolfe et al., 1987), and that insufficient time has passed to produce species-specific mtDNA mutations that might ultimately become fixed in each species. If this is correct, we might tentatively conclude that it is a faster mutation rate of ptDNA in A. chensiensis and A. fargesii rather than a higher rate of gene flow that caused the development of many rare species-specific ptDNA haplotypes in these two species. Such a difference in mutation rate between the two types of cytoplasmic DNA might also be a cause of ptDNA variation being more species specific in other conifer species pairs (groups) examined (see review by Du et al., 2009).
During the initial stages of divergence, it is to be expected that a large amount of neutral genetic variations would commonly be shared between closely related species, and that sequences with relatively faster mutation rates would become species specific more rapidly (de Queiroz, 2007). The replacement of shared ancestral genotypes by new species-specific genotypes would be accelerated by high rates of intraspecific gene flow enabling new mutant genotypes to expand rapidly throughout a species' range (Hoelzer, 1997). Thus, for older species pairs (groups) that originated and diverged from each other long ago, it would be expected that genetic markers showing relatively higher rates of mutation and gene flow should be more species specific and more likely to be fixed or occur at high frequency across a species' range (Zhou et al., 2010). In contrast, in very recently originated species pairs, such as the two fir species examined here, there would have been insufficient time for lineage sorting of ancestral polymorphisms to have been completed or for new markers originating by mutation to have spread widely and become fixed in a species.
It is clear from the comparative studies undertaken to date (Du et al., 2009, Zhou et al., 2010, and the present one), plus the results of recent modeling simulations (Currat et al., 2008), that different patterns of neutral genetic variation can exist within and between closely related species due to different rates of mutation and gene flow, and the length of time that has passed since speciation occurred. These factors are clearly of crucial importance when assessing how effective different genetic markers might be for delimiting species in particular groups.
Acknowledgments
This research was supported by grants from the Natural Science Foundation of China (30725004) and a Royal Society-NSF China International Joint Project award 2010/R4 to RJA and JQL.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abbott RJ, Smith LC, Milne RI, Crawford RM, Wolff K, Balfour J. Molecular analysis of plant migration and refugia in the Arctic. Science. 2000;289:1343–1346. doi: 10.1126/science.289.5483.1343. [DOI] [PubMed] [Google Scholar]
- Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, et al. Population history and natural selection shape patterns of genetic variation in 132 genes. PloS Biol. 2004;2:1591–1599. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Walker D. Pleistocene phylogeographical effects on avian populations and the speciation process. Philos Trans R Soc Lond B Biol Sci. 1998;265:457–463. doi: 10.1098/rspb.1998.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Brown GR, Gill GP, Kuntz RJ, Langley CH, Neale DB. Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc Natl Acad Sci USA. 2004;101:15255–15260. doi: 10.1073/pnas.0404231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KM, Abbott RJ, Milne RI, Tian XM, Liu JQ. Phylogeography of Pinus tabulaeformis Carr. (Pinaceae), a dominant species of coniferous forest in northern China. Mol Ecol. 2008;17:4276–4288. doi: 10.1111/j.1365-294x.2008.03911.x. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Templeton AR. Empirical tests of some predictions from coalescent theory with applications to intraspecic phylogeny reconstruction. Genetics. 1993;134:959–969. doi: 10.1093/genetics/134.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currat M, Manuel R, Petit JR, Excoffier L. The hidden side of invasion: massive introgression by local genes. Evolution. 2008;62:1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- de Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Demesure B, Sodzi N, Petit RJ. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Du KF, Petit RJ, Liu JQ. More introgression with less gene flow: chloroplast vs. mitochondrial DNA in the Picea asperata complex in china, and comparison with other conifers. Mol Ecol. 2009;18:1396–1407. doi: 10.1111/j.1365-294X.2009.04107.x. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol Ecol. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distance among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjon A, Rushforth KD. A classification of Abies Miller (Pinaceae) Notes R Bot Gard Edinburgh. 1989;46:59–77. [Google Scholar]
- Fu LG, Li N, Elias TS.1999PinaceaeIn: Wu Z, Raven PH (eds).Flora of ChinaVol. 4. Science Press: Beijing; 15–90. [Google Scholar]
- Gao LM, MöLLER M, Zhang XM, Hollingsworth ML, Liu J, Mill RR, et al. High variation and strong phylogeographic pattern among cpDNA haplotypes in Taxus wallichiana (Taxaceae) in China and North Vietnam. Mol Ecol. 2007;16:4684–4698. doi: 10.1111/j.1365-294X.2007.03537.x. [DOI] [PubMed] [Google Scholar]
- Gernandt DS, Magalión S, López GG, Flores OZ, Willyard A, Liston A. Use of simultaneous analyses to guide fossil-based calibrations of Pinaceae phylogeny. Int J Plant Sci. 2008;169:1086–1099. [Google Scholar]
- Godbout J, Fazekas A, Hewton CH, Yeh FC, Bousquet J. Glacial vicariance in the Pacific Northwest: evidence from a lodgepole pine mitochondrial DNA minisatellite for multiple genetically distinct and widely separated refugia. Mol Ecol. 2008;17:2463–2475. doi: 10.1111/j.1365-294X.2008.03761.x. [DOI] [PubMed] [Google Scholar]
- Goudet J.2001FSTAT (version 2.9.3): a program to estimate and test gene diversities and fixation indicesAvailable from http://www.unil.ch/izea/softwares/fstat.html .
- Gugger PF, Sugita S, Cavender-Bares J. Phylogeography of Douglas-fir based on mitochondrial and chloroplast DNA sequences: testing hypotheses from the fossil record. Mol Ecol. 2010;19:1877–1897. doi: 10.1111/j.1365-294X.2010.04622.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hoelzer GA. Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees revisited. Evolution. 1997;51:622–626. doi: 10.1111/j.1558-5646.1997.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hwang SY, Lin TP, Ma CS, Lin CL, Jeng-Der Chung, Yang JC. Postglacial population growth of Cunninghamia konishii (Cupressaceae) inferred from phylogeographical and mismatch analysis of chloroplast DNA variation. Mol Ecol. 2003;12:2689–2695. doi: 10.1046/j.1365-294x.2003.01935.x. [DOI] [PubMed] [Google Scholar]
- Jaramillo-correa JP, Beaulieu J, Bousquet J. Variation in mitochondrial DNA reveals multiple distant glacial refugial in black spruce (Picea mariana) a transcontinental North American conifer. Mol Ecol. 2004;13:2735–2747. doi: 10.1111/j.1365-294X.2004.02258.x. [DOI] [PubMed] [Google Scholar]
- Jaramillo-Correa JP, Aguirre-Planter E, Khasa DP, Eguiarte LE, Pinero D, Furnier GR, et al. Ancestry and divergence of subtropical montane forest isolates: molecular biogeography of the genus Abies (Pinaceae) in southern Mexico and Guatemala. Mol Ecol. 2008;17:2476–2490. doi: 10.1111/j.1365-294X.2008.03762.x. [DOI] [PubMed] [Google Scholar]
- Li SH, Yeung CK, Feinstein J, Han LX, Le MH, Wang CX, et al. Sailing through the Late Pleistocene: unusual historical demography of an East Asian endemic, the Chinese Hwamei (Leucodioptron canorum canorum), during the last glacial period. Mol Ecol. 2009;18:622–633. doi: 10.1111/j.1365-294X.2008.04028.x. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liepelt S, Bialozyt R, Ziegenhagen B. Wind-dispersed pollen mediates postglacial gene flow among refugia. Proc Natl Acad Sci USA. 2002;99:14590–14594. doi: 10.1073/pnas.212285399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepelt S, Mayland-Quellhorst E, Lahme M, Ziegenhagen B. Contrasting geographical patterns of ancient and modern genetic lineages in Mediterranean Abies species. Plant Syst Evol. 2010;284:141–151. [Google Scholar]
- Liu TS. A Monograph of the Genus Abies. Department of Forestry, College of Agriculture, National Taiwan University: Taipei; 1971. [Google Scholar]
- McKinnon GE, Jordan GJ, Vaillancourt RE, Steane DA, Potts BM. Glacial refugia and reticulate evolution: the case of the Tasmanian eucalypts. Philos Trans R Soc Lond B Biol Sci. 2004;359:275–284. doi: 10.1098/rstb.2003.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 2006;60:2399–2402. [PubMed] [Google Scholar]
- Palmé AE, Su Q, Palsson S, Lascoux M. Extensive sharing of chloroplast haplotypes among European birches indicates hybridization among Betula pendula, B. pubescens and B. nana. Mol Ecol. 2004;13:167–178. doi: 10.1046/j.1365-294x.2003.02034.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, et al. Glacial refugia: hotspot but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Excoffier L. Gene flow and species delimitation. Trends Ecol Evol. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Pons O, Petit RJ. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Qian H, Ricklefs RE. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature. 2000;407:180–182. doi: 10.1038/35025052. [DOI] [PubMed] [Google Scholar]
- Qiu YX, Guan BC, Fu CX, Comes HP. Did glacials and/or interglacials promote allopatric incipient speciation in East Asian temperate plants? Phylogeographic and coalescent analyses on refugial isolation and divergence in Dysosma versipellis. Mol Phylogenet Evol. 2009;51:281–293. doi: 10.1016/j.ympev.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence based phylogenetic analyses. Syst Biol. 2000;49:369–381. [PubMed] [Google Scholar]
- Sloan DB, Camille MB, Matthew SO, Stephen RK, Douglas RT. Evolutionary rate variation at multiple levels of biological organization in plant mitochondrial DNA. Mol Biol Evol. 2008;25:243–246. doi: 10.1093/molbev/msm266. [DOI] [PubMed] [Google Scholar]
- Swofford DL.2002PAUP*: Phylogenetic Analyses Using Parsimony (* and Other Methods)Version 4Sinauer Associates: Sunderland, MA [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Liu RR, Wang LY, Qiu Q, Chen KM, Liu JQ. Phylogeographic analyses suggest that a deciduous species (Ostryopsis davidiana Decne., Betulaceae) survived in northern China during the last glacial maximum. J Biogeogr. 2009;36:2148–2155. [Google Scholar]
- Ting CT, Tsaur SC, Wu CI. The phylogeny of closely related species as revealed by the genealogy of a speciation gene. Proc Natl Acad Sci USA. 2000;97:5313–5316. doi: 10.1073/pnas.090541597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura Y, Suyama Y. Differentiation of mitochondrial DNA polymorphisms in populations of five Japanese Abies species. Evolution. 1998;52:1031–1042. doi: 10.1111/j.1558-5646.1998.tb01831.x. [DOI] [PubMed] [Google Scholar]
- Via S. Natural selection in action during speciation. Proc Natl Acad Sci USA. 2009;106:9939–9946. doi: 10.1073/pnas.0901397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gao PX, Kang M, Lowe AJ, Huang HW. Refugia within refugia: the case study of a canopy tree (Eurycorymbus cavaleriei) in subtropical China. J Biogeogr. 2009;36:2156–2164. [Google Scholar]
- Wiens JJ. Species delimitation: new approaches for discovering diversity. Syst Biol. 2007;56:875–878. doi: 10.1080/10635150701748506. [DOI] [PubMed] [Google Scholar]
- Willyard A, Syring J, Gernandt DS, Liston A, Cronn R. Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations for Pinus. Mol Biol Evol. 2007;24:90–101. doi: 10.1093/molbev/msl131. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li W-H, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Krutovskii KV, Strauss SH. Abundant mitochondrial genome diversity, population differentiation and convergent evolution in pines. Genetics. 1998;150:1605–1614. doi: 10.1093/genetics/150.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LL, Cui XK, Milne RI, Sun YS, Liu JQ. Mutiple autopolyploidisations and range expansion of Allium przewalskianum Regel. (Allianceae) in the Qinghai-Tibetan Plateau. Mol Ecol. 2010;19:1691–1704. doi: 10.1111/j.1365-294X.2010.04613.x. [DOI] [PubMed] [Google Scholar]
- Xiang QP, Xiang QY, Guo YY, Zhang XC. Phylogeny of Abies (Pinaceae) inferred from nrITS sequence data. Taxon. 2009;58:141–152. [Google Scholar]
- Yi CL, Cui ZJ, Xiong HG. Numerical periods of Quaternary glaciations in China. Quatern Sci. 2005;25:609–619. [Google Scholar]
- Zheng Z. Vegetation and climate since the Late Pleistocene in southern China. J Geosci China. 2000;2:7–20. [Google Scholar]
- Zhou YF, Abbott RJ, Jiang ZY, Du FK, Milne RI, Liu JQ. Gene flow and species delimitation: a case study of two pine species with overlapping distributions in southeast China. Evolution. 2010;8:6342–6352. doi: 10.1111/j.1558-5646.2010.00988.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.