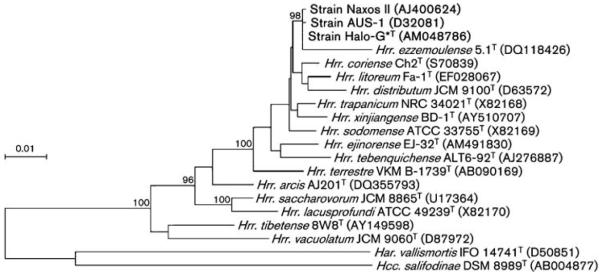
Fig. 1.
Phylogenetic tree based on the neighbour-joining algorithm, showing the relationships of strains Halo-G*T, AUS-1 and Naxos II and several Halorubrum type strains. The tree is based on an alignment of 16S rRNA gene sequences. Sequence accession numbers are given in parentheses. Bootstrap values higher than 80 out of 100 subreplicates are indicated at the respective bifurcations. The sequences of Haloarcula vallismortis IFO 14741T and Halococcus salifodinae DSM 8989T were used as the outgroup. Bar, 0.01 expected changes per site.